Interdependence of activation at rhaSR by cyclic AMP receptor protein, the RNA polymerase alpha subunit C-terminal domain, and rhaR - PubMed (original) (raw)
Interdependence of activation at rhaSR by cyclic AMP receptor protein, the RNA polymerase alpha subunit C-terminal domain, and rhaR
C C Holcroft et al. J Bacteriol. 2000 Dec.
Abstract
The Escherichia coli rhaSR operon encodes two AraC family transcription activators, RhaS and RhaR, and is activated by RhaR in the presence of L-rhamnose. beta-Galactosidase assays of various rhaS-lacZ promoter fusions combined with mobility shift assays indicated that a cyclic AMP receptor protein (CRP) site located at -111.5 is also required for full activation of rhaSR expression. To address the mechanisms of activation by CRP and the RNA polymerase alpha-subunit C-terminal domain (alpha-CTD) at rhaSR, we tested the effects of alanine substitutions in CRP activating regions 1 and 2, overexpression of a truncated version of alpha (alpha-Delta235), and alanine substitutions throughout alpha-CTD. We found that DNA-contacting residues in alpha-CTD are required for full activation, and for simplicity, we discuss alpha-CTD as a third activator of rhaSR. CRP and RhaR could each partially activate transcription in the absence of the other two activators, and alpha-CTD was not capable of activation alone. In the case of CRP, this suggests that this activation involves neither an alpha-CTD interaction nor cooperative binding with RhaR, while in the case of RhaR, this suggests the likelihood of direct interactions with core RNA polymerase. We also found that CRP, RhaR, and alpha-CTD each have synergistic effects on activation by the others, suggesting direct or indirect interactions among all three. We have some evidence that the alpha-CTD-CRP and alpha-CTD-RhaR interactions might be direct. The magnitude of the synergistic effects was usually greater with just two activators than with all three, suggesting possible redundancies in the mechanisms of activation by CRP, alpha-CTD, and RhaR.
Figures
FIG. 1
rhaSR-rhaBAD intergenic region. (A) Schematic representation of the rhaSR-rhaBAD intergenic region. The relative positions of the two RNA polymerases and the activator proteins RhaS, CRP, and RhaR are shown, as are the locations of the three putative CRP binding sites identified in this work. The activators and sites shown above the line all are located on one face of the DNA, and the activators and sites shown below the line are located on the opposite face. (B) The DNA sequence between the rhaBAD and rhaSR transcription start sites. The positions of the RhaS and RhaR binding sites are shown by everted arrows, and the positions of the CRP binding sites are shown as inverted arrows. The −10 and −35 hexamers of the two promoters are marked. Deletion endpoints (marked Δ), binding sites, and distances relative to the rhaBAD promoter are shown above the line, and deletion endpoints, binding sites, and distances relative to the rhaSR promoter are shown below the line. (C) Comparison of the putative CRP binding sites within the rhaSR-rhaBAD intergenic region and the CRP consensus binding site sequence. Nucleotides highlighted in gray match the consensus sequence.
FIG. 2
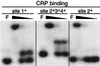
DNA mobility shift assays of CRP binding to sites 1, 2, 3, and 4. The fragment containing CRP site 1+ was PCR amplified using primers 742 and 744. The upstream primer 896 was used to generate fragments with sites 2+3+4+ and site 2+, with primers 1170 and 2165 used as downstream primers, respectively. Primers 742 and 896 were 32P labeled. The major band in the second lane in each set is at the position of the wells. Approximately 1 ng of 32P-labeled DNA fragment was added to each reaction mixture. The approximate CRP concentrations per reaction were the following: for the first lane in each set, F, none; for the second lane in each set, 8.4 μM CRP; for the third lane in each set, 2.1 μM CRP; and for the fourth lane in each set, 0.21 μM CRP.
FIG. 3
Effects of α-CTD alanine substitution mutants on rhaSR activation. Expression was measured from Φ(rhaS-lacZ)Δ_216_ (SME1074) cells carrying wild-type (w.t.) rpoA on a plasmid or a plasmid encoding α with a single alanine substitution at each position in α-CTD. Cells were grown in MOPS growth media containing glycerol,
l
-rhamnose, and 125 μg of ampicillin/ml. Values are the average of at least three independent assays and are shown as a percentage of the average expression from cells carrying wild-type rpoA on a plasmid. Analysis of variance was used to determine which alanine substitution mutants had significantly lower levels of expression compared to the wild type, which are indicated by an asterisk above the bar. β-Gal Act., β-galactosidase activity.
FIG. 4
Space-filling model of the predicted α-CTD structure, with residues that were defective at rhaSR highlighted. The model is based on the atomic coordinates of Jeon et al. (15). Colored residues are those identified as important at the Φ(rhaS-lacZ)Δ_216_ promoter fusion. Pink residues are those that may be involved in interaction with DNA, and green residues are those that have some other role, possibly protein-protein interactions. Residue numbers for some of the important residues are shown. The two models are related to one another by a 90° rotation on the vertical axis.
Similar articles
- Transcription activation by the DNA-binding domain of the AraC family protein RhaS in the absence of its effector-binding domain.
Wickstrum JR, Skredenske JM, Kolin A, Jin DJ, Fang J, Egan SM. Wickstrum JR, et al. J Bacteriol. 2007 Jul;189(14):4984-93. doi: 10.1128/JB.00530-07. Epub 2007 May 18. J Bacteriol. 2007. PMID: 17513476 Free PMC article. - Cyclic AMP receptor protein and RhaR synergistically activate transcription from the L-rhamnose-responsive rhaSR promoter in Escherichia coli.
Wickstrum JR, Santangelo TJ, Egan SM. Wickstrum JR, et al. J Bacteriol. 2005 Oct;187(19):6708-18. doi: 10.1128/JB.187.19.6708-6718.2005. J Bacteriol. 2005. PMID: 16166533 Free PMC article. - The AraC/XylS family activator RhaS negatively autoregulates rhaSR expression by preventing cyclic AMP receptor protein activation.
Wickstrum JR, Skredenske JM, Balasubramaniam V, Jones K, Egan SM. Wickstrum JR, et al. J Bacteriol. 2010 Jan;192(1):225-32. doi: 10.1128/JB.00829-08. J Bacteriol. 2010. PMID: 19854903 Free PMC article. - Protein-protein interactions during transcription activation: the case of the Escherichia coli cyclic AMP receptor protein.
Savery N, Rhodius V, Busby S. Savery N, et al. Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):543-50. doi: 10.1098/rstb.1996.0053. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735277 Review. - Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme.
McFall SM, Chugani SA, Chakrabarty AM. McFall SM, et al. Gene. 1998 Nov 26;223(1-2):257-67. doi: 10.1016/s0378-1119(98)00366-7. Gene. 1998. PMID: 9858745 Review.
Cited by
- Amino acid contacts between sigma 70 domain 4 and the transcription activators RhaS and RhaR.
Wickstrum JR, Egan SM. Wickstrum JR, et al. J Bacteriol. 2004 Sep;186(18):6277-85. doi: 10.1128/JB.186.18.6277-6285.2004. J Bacteriol. 2004. PMID: 15342598 Free PMC article. - Growing repertoire of AraC/XylS activators.
Egan SM. Egan SM. J Bacteriol. 2002 Oct;184(20):5529-32. doi: 10.1128/JB.184.20.5529-5532.2002. J Bacteriol. 2002. PMID: 12270809 Free PMC article. Review. No abstract available. - Transcription activation by the DNA-binding domain of the AraC family protein RhaS in the absence of its effector-binding domain.
Wickstrum JR, Skredenske JM, Kolin A, Jin DJ, Fang J, Egan SM. Wickstrum JR, et al. J Bacteriol. 2007 Jul;189(14):4984-93. doi: 10.1128/JB.00530-07. Epub 2007 May 18. J Bacteriol. 2007. PMID: 17513476 Free PMC article. - Differences in the mechanism of the allosteric l-rhamnose responses of the AraC/XylS family transcription activators RhaS and RhaR.
Kolin A, Balasubramaniam V, Skredenske JM, Wickstrum JR, Egan SM. Kolin A, et al. Mol Microbiol. 2008 Apr;68(2):448-61. doi: 10.1111/j.1365-2958.2008.06164.x. Mol Microbiol. 2008. PMID: 18366439 Free PMC article. - Mechanism of ToxT-dependent transcriptional activation at the Vibrio cholerae tcpA promoter.
Hulbert RR, Taylor RK. Hulbert RR, et al. J Bacteriol. 2002 Oct;184(20):5533-44. doi: 10.1128/JB.184.20.5533-5544.2002. J Bacteriol. 2002. PMID: 12270810 Free PMC article.
References
- Berg O G, von Hippel P H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988;200:709–723. - PubMed
- Blatter E E, Ross W, Tang H, Gourse R L, Ebright R H. Domain organization of RNA polymerase α subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell. 1994;78:889–896. - PubMed
- Busby S, Ebright R H. Transcription activation at class II CAP-dependent promoters. Mol Microbiol. 1997;23:853–859. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous