Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells - PubMed (original) (raw)
Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells
C Richardson et al. Mol Cell Biol. 2000 Dec.
Abstract
DNA double-strand breaks (DSBs) may be caused by normal metabolic processes or exogenous DNA damaging agents and can promote chromosomal rearrangements, including translocations, deletions, or chromosome loss. In mammalian cells, both homologous recombination and nonhomologous end joining (NHEJ) are important DSB repair pathways for the maintenance of genomic stability. Using a mouse embryonic stem cell system, we previously demonstrated that a DSB in one chromosome can be repaired by recombination with a homologous sequence on a heterologous chromosome, without any evidence of genome rearrangements (C. Richardson, M. E. Moynahan, and M. Jasin, Genes Dev., 12:3831-3842, 1998). To determine if genomic integrity would be compromised if homology were constrained, we have now examined interchromosomal recombination between truncated but overlapping gene sequences. Despite these constraints, recombinants were readily recovered when a DSB was introduced into one of the sequences. The overwhelming majority of recombinants showed no evidence of chromosomal rearrangements. Instead, events were initiated by homologous invasion of one chromosome end and completed by NHEJ to the other chromosome end, which remained highly preserved throughout the process. Thus, genomic integrity was maintained by a coupling of homologous and nonhomologous repair pathways. Interestingly, the recombination frequency, although not the structure of the recombinant repair products, was sensitive to the relative orientation of the gene sequences on the interacting chromosomes.
Figures
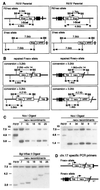
FIG. 1
Interchromosomal recombination substrates. The 5′neo allele (A) was constructed by targeting 5′neo and the hyg gene to the pim-1 locus on chr.17. The forward orientation (F allele) of the 5′neo sequence is present in the F5′/3′ cell lines, and the reverse orientation (R allele) is present in the R5′/3′ cell lines, as indicated by the arrow orientation. The 3′neo allele (B) was constructed by targeting 3′neo and the HPRT gene to the Rb locus on chr.14. Wild-type (top) and targeted (bottom) alleles are diagrammed for each locus, with confirmatory Southern analyses presented using _Hin_cII and _Pst_I for the 5′neo and the 3′neo alleles, respectively, and probing with sequences as indicated. An I-_Sce_I site disrupts the 5′neo gene at the _Nco_I site. Two independently derived clones were analyzed for both F5′/3′ and R5′/3′, as indicated (F5′/3′, F12 and G12; R5′/3′, B3 and C11). Black bars, pim-1 (A) or Rb (B) exons. HII, _Hin_cII; Pst, _Pst_I; Nco, _Nco_I. (C) The truncated 5′neo and 3′neo sequences. Distances are indicated in bp. 5′neo contains a promoter, and 3′neo contains the neo gene stop codon and a polyadenylation signal. There is 468 bp of overlap between the 5′neo and 3′neo sequences.
FIG. 2
DSB repair products. Restriction maps of parental (A) and repaired neo+ alleles (B). _Nco_I digestion is diagnostic for homologous repair of the I-_Sce_I-generated DSB in either 5′neo allele. _Bgl_II/_Sac_II digestion is used to characterize the type of recombination event. _Bgl_II/_Sac_II fragments of variable size (B) indicate gene conversion tracts incorporating variable lengths of chr.14 sequences (+chr.14) and NHEJ at variable positions of chr.17. BII, _Bgl_II; N, _Nco_I; SII, _Sac_II. (C) Southern blot analysis of representative clones. Note that the 3′neo allele is unchanged from the parental allele in each of the recombinant clones. The probe was a 5′neo gene fragment. (D) Position of PCR primers derived from chr.17 sequences that flank the repair junctions.
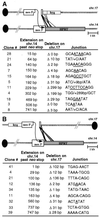
FIG. 3
Sequence analysis of repair junctions from class II neo+ clones. PCR products from the neo+ allele of the F_neo_+/3′ (A) and R_neo_+/3′ cell lines (B) were cloned and sequenced. Extension on chr.14 indicates the length of newly synthesized DNA incorporated at one end of the chr.17 break site beyond the neo stop codon. Thus, the total extension includes an additional 229 bp to the number indicated. Deletion on chr.17 indicates the number of base pairs deleted from the nonconverted end of the I-_Sce_I break site prior to NHEJ. The sequences at the junctions of chr.14 and chr.17 are separated by a hyphen when there is no sequence overlap, underlined if microhomology is present, or indicated by the base pairs that were inserted at the junction.
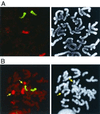
FIG. 4
Visualization of a rare translocation by FISH using whole chromosome mouse chr.14 fluorescein isothiocyanate (green) and chr.17 Cy3 (red) probes (left panel) and DAPI (right panel). (A) Representative class IV clone having two normal chrs.14 and chrs.17. (B) Reciprocal translocation clone (R_neo_+/3′ clone 17) having two normal chrs.14, one normal untargeted chr.17, and two hybrid translocation chromosomes involving chr.17 and an unidentified chromosome (red/unstained), as indicated by the arrows.
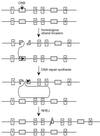
FIG. 5
Model for partial tandem gene duplications. Events can be initiated by a DSB within a repetitive element (gray box) on one chromosome, followed by invasion of one end into the same element on the homologue or sister chromatid. Repair synthesis extends downstream to duplicate sequences, shown here as exons 2 and 3 (white bars). Completion of the repair event occurs by NHEJ of the newly synthesized strands at or near the end of the broken chromosome. The result is a partial tandem gene duplication on one chromosome or chromatid, while the other chromosome or chromatid remains unaltered.
Similar articles
- Analysis of DNA double-strand break repair pathways in mice.
Brugmans L, Kanaar R, Essers J. Brugmans L, et al. Mutat Res. 2007 Jan 3;614(1-2):95-108. doi: 10.1016/j.mrfmmm.2006.01.022. Epub 2006 Jun 23. Mutat Res. 2007. PMID: 16797606 Review. - Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks.
Stark JM, Jasin M. Stark JM, et al. Mol Cell Biol. 2003 Jan;23(2):733-43. doi: 10.1128/MCB.23.2.733-743.2003. Mol Cell Biol. 2003. PMID: 12509470 Free PMC article. - Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations.
Richardson C, Moynahan ME, Jasin M. Richardson C, et al. Genes Dev. 1998 Dec 15;12(24):3831-42. doi: 10.1101/gad.12.24.3831. Genes Dev. 1998. PMID: 9869637 Free PMC article. - Radiation-induced genomic rearrangements formed by nonhomologous end-joining of DNA double-strand breaks.
Rothkamm K, Kühne M, Jeggo PA, Löbrich M. Rothkamm K, et al. Cancer Res. 2001 May 15;61(10):3886-93. Cancer Res. 2001. PMID: 11358801
Cited by
- Role of non-homologous end joining in V(D)J recombination.
Malu S, Malshetty V, Francis D, Cortes P. Malu S, et al. Immunol Res. 2012 Dec;54(1-3):233-46. doi: 10.1007/s12026-012-8329-z. Immunol Res. 2012. PMID: 22569912 Review. - An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells.
Delacôte F, Han M, Stamato TD, Jasin M, Lopez BS. Delacôte F, et al. Nucleic Acids Res. 2002 Aug 1;30(15):3454-63. doi: 10.1093/nar/gkf452. Nucleic Acids Res. 2002. PMID: 12140331 Free PMC article. - Sequence conversion by single strand oligonucleotide donors via non-homologous end joining in mammalian cells.
Liu J, Majumdar A, Liu J, Thompson LH, Seidman MM. Liu J, et al. J Biol Chem. 2010 Jul 23;285(30):23198-207. doi: 10.1074/jbc.M110.123844. Epub 2010 May 19. J Biol Chem. 2010. PMID: 20489199 Free PMC article. - Increased sensitivity of subtelomeric regions to DNA double-strand breaks in a human cancer cell line.
Zschenker O, Kulkarni A, Miller D, Reynolds GE, Granger-Locatelli M, Pottier G, Sabatier L, Murnane JP. Zschenker O, et al. DNA Repair (Amst). 2009 Aug 6;8(8):886-900. doi: 10.1016/j.dnarep.2009.05.004. Epub 2009 Jun 18. DNA Repair (Amst). 2009. PMID: 19540174 Free PMC article. - Mechanisms driving chromosomal translocations: lost in time and space.
Ramsden DA, Nussenzweig A. Ramsden DA, et al. Oncogene. 2021 Jun;40(25):4263-4270. doi: 10.1038/s41388-021-01856-9. Epub 2021 Jun 8. Oncogene. 2021. PMID: 34103687 Free PMC article. Review.
References
- Belmaaza A, Chartrand P. One-sided invasion events in homologous recombination at double-strand breaks. Mutat Res. 1994;314:199–208. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources