Horizontal gene transfer in bacterial and archaeal complete genomes - PubMed (original) (raw)
Comparative Study
. 2000 Nov;10(11):1719-25.
doi: 10.1101/gr.130000.
Affiliations
- PMID: 11076857
- PMCID: PMC310969
- DOI: 10.1101/gr.130000
Comparative Study
Horizontal gene transfer in bacterial and archaeal complete genomes
S Garcia-Vallvé et al. Genome Res. 2000 Nov.
Abstract
There is growing evidence that horizontal gene transfer is a potent evolutionary force in prokaryotes, although exactly how potent is not known. We have developed a statistical procedure for predicting whether genes of a complete genome have been acquired by horizontal gene transfer. It is based on the analysis of G+C contents, codon usage, amino acid usage, and gene position. When we applied this procedure to 17 bacterial complete genomes and seven archaeal ones, we found that the percentage of horizontally transferred genes varied from 1.5% to 14.5%. Archaea and nonpathogenic bacteria had the highest percentages and pathogenic bacteria, except for Mycoplasma genitalium, had the lowest. As reported in the literature, we found that informational genes were less likely to be transferred than operational genes. Most of the horizontally transferred genes were only present in one or two lineages. Some of these transferred genes include genes that form part of prophages, pathogenecity islands, transposases, integrases, recombinases, genes present only in one of the two Helicobacter pylori strains, and regions of genes functionally related. All of these findings support the important role of horizontal gene transfer in the molecular evolution of microorganisms and speciation.
Figures
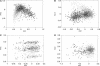
Figure 1
Correspondence analysis of relative synonymous usage for genes of (A) Escherichia coli, (B) Bacillus subtilis, (C) Thermotoga maritima, and (D) Methanobacterium thermoautotrophicum. Triangles correspond to genes proposed as being acquired by horizontal gene transfer and included in regions with a low G+C content. Circles correspond to genes proposed as being acquired by HGT and included in regions with a high G+C content. Dots correspond to genes that are not proposed as being acquired by HGT. For the sake of clarity, genes proposed as being acquired by HGT that do not belong to a region that is high or low in the G+C content are not shown.
Similar articles
- Hundreds of novel composite genes and chimeric genes with bacterial origins contributed to haloarchaeal evolution.
Méheust R, Watson AK, Lapointe FJ, Papke RT, Lopez P, Bapteste E. Méheust R, et al. Genome Biol. 2018 Jun 7;19(1):75. doi: 10.1186/s13059-018-1454-9. Genome Biol. 2018. PMID: 29880023 Free PMC article. - Evolution of mosaic operons by horizontal gene transfer and gene displacement in situ.
Omelchenko MV, Makarova KS, Wolf YI, Rogozin IB, Koonin EV. Omelchenko MV, et al. Genome Biol. 2003;4(9):R55. doi: 10.1186/gb-2003-4-9-r55. Epub 2003 Aug 29. Genome Biol. 2003. PMID: 12952534 Free PMC article. - Origins of major archaeal clades correspond to gene acquisitions from bacteria.
Nelson-Sathi S, Sousa FL, Roettger M, Lozada-Chávez N, Thiergart T, Janssen A, Bryant D, Landan G, Schönheit P, Siebers B, McInerney JO, Martin WF. Nelson-Sathi S, et al. Nature. 2015 Jan 1;517(7532):77-80. doi: 10.1038/nature13805. Epub 2014 Oct 15. Nature. 2015. PMID: 25317564 Free PMC article. - Archaeal integrative genetic elements and their impact on genome evolution.
She Q, Brügger K, Chen L. She Q, et al. Res Microbiol. 2002 Jul-Aug;153(6):325-32. doi: 10.1016/s0923-2508(02)01331-1. Res Microbiol. 2002. PMID: 12234006 Review. - Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world.
Koonin EV, Wolf YI. Koonin EV, et al. Nucleic Acids Res. 2008 Dec;36(21):6688-719. doi: 10.1093/nar/gkn668. Epub 2008 Oct 23. Nucleic Acids Res. 2008. PMID: 18948295 Free PMC article. Review.
Cited by
- Use of a multi-way method to analyze the amino acid composition of a conserved group of orthologous proteins in prokaryotes.
Pasamontes A, Garcia-Vallve S. Pasamontes A, et al. BMC Bioinformatics. 2006 May 18;7:257. doi: 10.1186/1471-2105-7-257. BMC Bioinformatics. 2006. PMID: 16709240 Free PMC article. - Whole genome evaluation of horizontal transfers in the pathogenic fungus Aspergillus fumigatus.
Mallet LV, Becq J, Deschavanne P. Mallet LV, et al. BMC Genomics. 2010 Mar 12;11:171. doi: 10.1186/1471-2164-11-171. BMC Genomics. 2010. PMID: 20226043 Free PMC article. - Horizontal transfer of two operons coding for hydrogenases between bacteria and archaea.
Calteau A, Gouy M, Perrière G. Calteau A, et al. J Mol Evol. 2005 May;60(5):557-65. doi: 10.1007/s00239-004-0094-8. J Mol Evol. 2005. PMID: 15983865 - Inferring genome trees by using a filter to eliminate phylogenetically discordant sequences and a distance matrix based on mean normalized BLASTP scores.
Clarke GD, Beiko RG, Ragan MA, Charlebois RL. Clarke GD, et al. J Bacteriol. 2002 Apr;184(8):2072-80. doi: 10.1128/JB.184.8.2072-2080.2002. J Bacteriol. 2002. PMID: 11914337 Free PMC article. - Exploration of the Biosynthetic Potential of the Populus Microbiome.
Blair PM, Land ML, Piatek MJ, Jacobson DA, Lu TS, Doktycz MJ, Pelletier DA. Blair PM, et al. mSystems. 2018 Oct 2;3(5):e00045-18. doi: 10.1128/mSystems.00045-18. eCollection 2018 Sep-Oct. mSystems. 2018. PMID: 30320216 Free PMC article.
References
- Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. - PubMed
- Chou KC, Zhang CT. Prediction of protein structural classes. Crit Rev Biochem Mol Biol. 1995;30:275–349. - PubMed
- Doolittle RF. Microbial genomes opened up. Nature. 1998;392:339–342. - PubMed
- Doolittle WF. Lateral genomics. Trends Biochem Sci. 1999a;24:M5–M8. - PubMed
- ————— Phylogenetic classification and the universal tree. Science. 1999b;284:2124–2129. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials