DNA cloning using in vitro site-specific recombination - PubMed (original) (raw)
DNA cloning using in vitro site-specific recombination
J L Hartley et al. Genome Res. 2000 Nov.
Abstract
As a result of numerous genome sequencing projects, large numbers of candidate open reading frames are being identified, many of which have no known function. Analysis of these genes typically involves the transfer of DNA segments into a variety of vector backgrounds for protein expression and functional analysis. We describe a method called recombinational cloning that uses in vitro site-specific recombination to accomplish the directional cloning of PCR products and the subsequent automatic subcloning of the DNA segment into new vector backbones at high efficiency. Numerous DNA segments can be transferred in parallel into many different vector backgrounds, providing an approach to high-throughput, in-depth functional analysis of genes and rapid optimization of protein expression. The resulting subclones maintain orientation and reading frame register, allowing amino- and carboxy-terminal translation fusions to be generated. In this paper, we outline the concepts of this approach and provide several examples that highlight some of its potential.
Figures
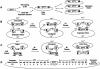
Figure 1
(A) Schematic of gene cloning and transfer by recombinational cloning. The triangles represent recombination sites. Genes are cloned into Entry Vectors by in vitro recombination of PCR products or with restriction enzymes and ligase. Thereafter, genes are moved entirely by recombination. (B) Subcloning of a gene from the Entry Vector into a Destination Vector. (C) Cloning _att_B-PCR products by in vitro recombination. (D) Sequences of the _att_B1 and _att_B2 sites that flank the gene either in a PCR product made with primers containing _att_B sites or in an Expression Clone.
Figure 2
(A) Agarose gel (1%) of PCR amplification products (2.5 μL of 35 μL) using _att_B-primers (see Methods) encoding eIF4e (lane 1), tyrosine kinase (lane 2), transferrin receptor (lane 3), β-adaptin (lane 4), MAP4 (lane 5), glucuronidase (Gus, lane 6), or the tetracycline-resistance gene (_Tet_R; lane 7). (B) _att_P cloning vector pDONR203. PCR products cloned by in vitro recombination replace the chloramphenicol-resistance and _ccd_B genes; the recombination reactions convert the _att_P sites to _att_L sites. (C) Colonies resulting from transformation of E. coli with reactions (2 μL of 22 μL) containing the PCR products, pDONR203, and BP Clonase. The negative control contained all components except PCR product. (D) Entry Clone pENTR203-eIF4e, the product of recombinational cloning of the eIF4e PCR product into pDONR203. (E) Miniprep DNA (Entry Clones) from the colonies in C. Lanes 1_–_4, eIF4e; lanes 5_–_8, tyrosine kinase; lanes 9_–_12, transferrin receptor; lanes 13_–_16, β-adaptin; lanes 17_–_20, MAP4; lanes 21_–_24, Gus; M, supercoiled DNA ladder.
Figure 3
(A) Destination Vector pDEST17, for expressing His6 fusions in Escherichia coli. (B) Colonies resulting from transformation of DH5α cells with 2 μL (of 22 μL) of LR reactions. The negative control contained all components except the Entry Clone DNA. pDEST17 was linearized at the unique NcoI site prior to mixing with Entry Clone DNA. (C) The Expression (_att_B) clone pEXP17-eIF4e, which resulted from the LR reaction with pENTR203-eIF4e and pDEST17. The subcloned eIF4e gene has replaced the chloramphenicol resistance-_ccd_B segment, with the amino end of the gene downstream of the T7 promoter and the His6 tag in frame with the eIF4e open reading frame. The recombination reactions convert the _att_R sites to _att_B sites (25 bp). (D) Miniprep DNA of single colonies (Expression Clones) from each reaction. Lane 1, eIF4e; lane 2, tyrosine kinase; lane 3, transferrin receptor; lane 4, β-adaptin; lane 5, MAP4; lane 6, Gus; lane 7; _Tet_R. M, supercoiled DNA ladder.
Figure 4
(A) SDS-PAGE gel of proteins expressed from Expression Clones of the genes in pDEST17 (His6 fusion) vector in Escherichia coli strain BL21 SI. (Lane 1), eIF4e; (lane 2), tyrosine kinase; (lane 3), transferrin receptor; (lane 4), β-adaptin; (lane 5), MAP4 (MAP4 is known to migrate aberrantly on SDS-PAGE gels; Chapin et al. 1995); (lane 6), Gus. (B) SDS-PAGE gel of expression of genes subcloned from the same Entry Clones as in (A) into baculovirus Destination Vectors. (Lanes 1_–_5) are pDest8 (native expression; (lane 1), eIF4e; (lane 2), tyrosine kinase; (lane 3), transferrin receptor; (lane 4), β-adaptin; (lane 5), MAP4). (Lanes 6_–_8) the gus gene, from the same Entry Clone as in A, expressed in the baculovirus vectors pDest8 (lane 6, native, 68 kD), pDest10 (lane 7, His6 fusion, 74 kD), and pDest20 (lane 8, GST fusion, 97 kD). As in E. coli expression (above), MAP4 protein migrated as a ∼200-kD protein instead of as a 121-kD protein (Chapin et al. 1995).
Similar articles
- The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes.
Liu Q, Li MZ, Leibham D, Cortez D, Elledge SJ. Liu Q, et al. Curr Biol. 1998 Dec 3;8(24):1300-9. doi: 10.1016/s0960-9822(07)00560-x. Curr Biol. 1998. PMID: 9843682 - Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones.
Cheo DL, Titus SA, Byrd DR, Hartley JL, Temple GF, Brasch MA. Cheo DL, et al. Genome Res. 2004 Oct;14(10B):2111-20. doi: 10.1101/gr.2512204. Genome Res. 2004. PMID: 15489333 Free PMC article. - From genes to proteins: in vitro expression of rickettsial proteins.
Renesto P, Raoult D. Renesto P, et al. Ann N Y Acad Sci. 2003 Jun;990:642-52. doi: 10.1111/j.1749-6632.2003.tb07439.x. Ann N Y Acad Sci. 2003. PMID: 12860702 Review. - Rapid one-step recombinational cloning.
Fu C, Wehr DR, Edwards J, Hauge B. Fu C, et al. Nucleic Acids Res. 2008 May;36(9):e54. doi: 10.1093/nar/gkn167. Epub 2008 Apr 19. Nucleic Acids Res. 2008. PMID: 18424799 Free PMC article. - High throughput cloning and expression strategies for protein production.
Betton JM. Betton JM. Biochimie. 2004 Sep-Oct;86(9-10):601-5. doi: 10.1016/j.biochi.2004.07.004. Biochimie. 2004. PMID: 15556269 Review.
Cited by
- Knockdown of ttc26 disrupts ciliogenesis of the photoreceptor cells and the pronephros in zebrafish.
Zhang Q, Liu Q, Austin C, Drummond I, Pierce EA. Zhang Q, et al. Mol Biol Cell. 2012 Aug;23(16):3069-78. doi: 10.1091/mbc.E12-01-0019. Epub 2012 Jun 20. Mol Biol Cell. 2012. PMID: 22718903 Free PMC article. - A pipeline for the systematic identification of non-redundant full-ORF cDNAs for polymorphic and evolutionary divergent genomes: Application to the ascidian Ciona intestinalis.
Gilchrist MJ, Sobral D, Khoueiry P, Daian F, Laporte B, Patrushev I, Matsumoto J, Dewar K, Hastings KE, Satou Y, Lemaire P, Rothbächer U. Gilchrist MJ, et al. Dev Biol. 2015 Aug 15;404(2):149-63. doi: 10.1016/j.ydbio.2015.05.014. Epub 2015 May 27. Dev Biol. 2015. PMID: 26025923 Free PMC article. - A role for APETALA1/fruitfull transcription factors in tomato leaf development.
Burko Y, Shleizer-Burko S, Yanai O, Shwartz I, Zelnik ID, Jacob-Hirsch J, Kela I, Eshed-Williams L, Ori N. Burko Y, et al. Plant Cell. 2013 Jun;25(6):2070-83. doi: 10.1105/tpc.113.113035. Epub 2013 Jun 14. Plant Cell. 2013. PMID: 23771895 Free PMC article. - Principles and Practical Considerations for the Analysis of Disease-Associated Alternative Splicing Events Using the Gateway Cloning-Based Minigene Vectors pDESTsplice and pSpliceExpress.
Putscher E, Hecker M, Fitzner B, Lorenz P, Zettl UK. Putscher E, et al. Int J Mol Sci. 2021 May 13;22(10):5154. doi: 10.3390/ijms22105154. Int J Mol Sci. 2021. PMID: 34068052 Free PMC article. Review. - Hematopoietic stem cell and gene therapy corrects primary neuropathology and behavior in mucopolysaccharidosis IIIA mice.
Langford-Smith A, Wilkinson FL, Langford-Smith KJ, Holley RJ, Sergijenko A, Howe SJ, Bennett WR, Jones SA, Wraith J, Merry CL, Wynn RF, Bigger BW. Langford-Smith A, et al. Mol Ther. 2012 Aug;20(8):1610-21. doi: 10.1038/mt.2012.82. Epub 2012 May 1. Mol Ther. 2012. PMID: 22547151 Free PMC article.
References
- Abremski K, Hoess R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J Biol Chem. 1984;259:1509–1514. - PubMed
- Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226:735–745. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials