Lava lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization - PubMed (original) (raw)
Lava lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization
J C Sisson et al. J Cell Biol. 2000.
Abstract
Drosophila cellularization and animal cell cytokinesis rely on the coordinated functions of the microfilament and microtubule cytoskeletal systems. To identify new proteins involved in cellularization and cytokinesis, we have conducted a biochemical screen for microfilament/microtubule-associated proteins (MMAPs). 17 MMAPs were identified; seven have been previously implicated in cellularization and/or cytokinesis, including KLP3A, Anillin, Septins, and Dynamin. We now show that a novel MMAP, Lava Lamp (Lva), is also required for cellularization. Lva is a coiled-coil protein and, unlike other proteins previously implicated in cellularization or cytokinesis, it is Golgi associated. Our functional analysis shows that cellularization is dramatically inhibited upon injecting anti-Lva antibodies (IgG and Fab) into embryos. In addition, we show that brefeldin A, a potent inhibitor of membrane trafficking, also inhibits cellularization. Biochemical analysis demonstrates that Lva physically interacts with the MMAPs Spectrin and CLIP190. We suggest that Lva and Spectrin may form a Golgi-based scaffold that mediates the interaction of Golgi bodies with microtubules and facilitates Golgi-derived membrane secretion required for the formation of furrows during cellularization. Our results are consistent with the idea that animal cell cytokinesis depends on both actomyosin-based contraction and Golgi-derived membrane secretion.
Figures
Figure 1
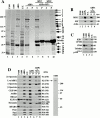
The purification of Drosophila MMAPs. (A) This Coomassie-stained SDS-polyacrylamide gradient gel shows the proteins within different fractions from the purification: high-speed pellet (P100) and supernatant (S100, column load), F-actin–binding proteins (ABPs), and F-actin column flow through (F.T.). Lanes 1–4 were each loaded with 25 μg of total protein. ABPs do not sediment without MTs (lanes 5 and 6), but a small subset of ABPs do sediment with MTs (spin 1). The lane 8 pellet was rinsed, resuspended, and the MTs were resedimented (spin 2). Numbered arrows indicate the proteins that resediment with the MTs. Comparable amounts of each matched supernatant (S) and pellet (P) were loaded. Molecular weight standards are indicated to the left. *Presumed protein degradation. (B) Immunoblots for MHC, 95F, and actin. Lanes are labeled and loaded as in A, lanes 5 and 6 correspond to spin 1 in A. (C) Immunoblots for the indicated MT-binding proteins, and tubulin. Each lane is labeled and loaded as in A. (D, left) Equivalent protein blots probed with antibodies to the indicated proteins. Each lane is labeled and loaded as in A. Coomassie-stained band numbers and molecular weights are indicated to the right. (Right) S100 (25 μg) protein blot probed with affinity-purified rabbit anti–Lva antibody. Molecular weight standards are indicated.
Figure 3
Lva associates with Spectrins and CLIP190. Immunoblots show (A) gel filtration elution profiles for the indicated proteins in the S100 and MMAP fractions. Fraction numbers and the elution peaks for native molecular weight standards are shown at the top. (B) Elution profiles for the indicated proteins that rebind an F-actin column. Lane headings indicate F-actin column load (ABPs), F-actin column flow through (F.T.), and elution fraction numbers. Identical results for each protein were also obtained with the MMAP fraction (data not shown). (C) A mock IP performed with BSA, and IPs performed with anti–Lva, anti–α-Spectrin, and anti–CLIP190 antibodies on the MMAP fraction. Proteins detected by immunoblot are indicated to the left. Comparable amounts of each matched supernatant (S) and pellet (P) were loaded.
Figure 2
Molecular characterization of Lva. (A) The predicted gene structure shows exons (black rectangles) and introns (gaps). Exons are numbered 1–8. The gene prediction analysis was performed with the program FGENESH (available online from Baylor College of Medicine, Waco, TX). The positions of the putative start codon (bent arrow), stop codon (straight arrow), and the 5′ extent of the cDNA sequence (vertical arrowhead) are as indicated. (B) Lva is predicted to be globular at its termini (shaded) and form a coiled-coil along its central region (black). Arrowheads indicate the junctions between amino acids (a.a.) encoded by adjacent exons and the black marks below the diagram indicate the position of peptides matching MS/MS data.
Figure 5
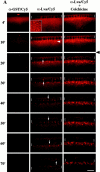
Golgi bodies undergo dynamic, MT-dependent movements in living embryos. (A) Frames from scanning confocal movies show three different Myo-GFP embryos injected at the start of cycle 14 with either diluted Cy5-tagged anti–GST (left), Cy5-tagged anti–Lva (center), or Cy5-tagged anti–Lva antibodies followed by injection of 25 mM colchicine 12.5 min into cycle 14 (right). The black arrowhead indicates the colchicine injection time. Brackets indicate the position of furrow fronts, arrows indicate the direction of Golgi movement (apical is up), and the white arrowhead indicates Cy5-marked Golgi bodies just basal of nuclei. Time (minutes) is relative to the start of cycle 14. The dorsal, injected surface of each embryo is shown. The inset shows a grazing view of pericentrosomal Golgi ∼1.0 μm below the cell surface. All other views are sagittal. Bar, 10 μm. (B) Sagittal view of a cellularizing embryo that was injected with dilute, Cy5-tagged anti–Lva antibody (middle), fixed, and counter stained with anti–Golgi p120 antibody (left) with merged panel (right). Bar, 5 μm.
Figure 5
Golgi bodies undergo dynamic, MT-dependent movements in living embryos. (A) Frames from scanning confocal movies show three different Myo-GFP embryos injected at the start of cycle 14 with either diluted Cy5-tagged anti–GST (left), Cy5-tagged anti–Lva (center), or Cy5-tagged anti–Lva antibodies followed by injection of 25 mM colchicine 12.5 min into cycle 14 (right). The black arrowhead indicates the colchicine injection time. Brackets indicate the position of furrow fronts, arrows indicate the direction of Golgi movement (apical is up), and the white arrowhead indicates Cy5-marked Golgi bodies just basal of nuclei. Time (minutes) is relative to the start of cycle 14. The dorsal, injected surface of each embryo is shown. The inset shows a grazing view of pericentrosomal Golgi ∼1.0 μm below the cell surface. All other views are sagittal. Bar, 10 μm. (B) Sagittal view of a cellularizing embryo that was injected with dilute, Cy5-tagged anti–Lva antibody (middle), fixed, and counter stained with anti–Golgi p120 antibody (left) with merged panel (right). Bar, 5 μm.
Figure 4
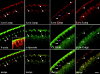
Lva, α-Spectrin, and CLIP190 colocalize to Golgi bodies. Cellularizing embryos were fixed, labeled with pairs of fluorescent probes, and examined by scanning confocal microscopy. From left to right, matched panels show Lva with F-actin, α-Spectrin, CLIP190, and the Golgi marker p120, respectively. White arrowheads indicate corresponding puncta in matched panels, except in the Lva/F-actin panels, where they indicate puncta at the furrow front. The purple arrowhead in the CLIP190 panel indicates CLIP190 with no corresponding Lva fluorescence. (Inset) Cortical F-actin (green) and MT-inverted baskets (red). Essentially no fluorescence was detected with secondary antibodies alone (data not shown). All views are sagittal. Bar, 5 μm.
Figure 6
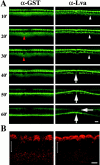
Anti–Lva IgG antibody inhibits furrow progression. All panels are from scanning confocal movies of Myo-GFP embryos. (A, left) Anti–GST antibody-injected embryo undergoing normal furrowing. The red arrowheads indicate an injection artifact that the embryo repairs. (Right) Anti–Lva IgG antibody (3.8 mg/ml)-injected embryo undergoing disrupted furrowing (vertical arrows). A surface dimple forms near the site of injection where furrowing is most impaired (horizontal arrow). The Myo-GFP puncta observed near the site of the anti–Lva antibody injection (white arrowheads) and the smaller Myo-GFP puncta observed normally in both embryos do not colocalize with anti–Lva/Cy5 antibody marked Golgi (data not shown). Time (minutes) is relative to the start of cycle 14. The dorsal, injected surface of each embryo is shown. All views are sagittal. Bar, 10 μm. (B) Sagittal views of two cellularizing embryos that were injected with either anti–GST (left) or anti–Lva (right) IgG antibodies, fixed, and stained with anti–Golgi p120 antibody. Brackets indicate the position of nuclei (apical is up). Bar, 5 μm.
Figure 7
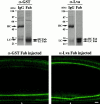
Anti–Lva Fab antibody inhibits furrow progression. (Top) Coomassie stained 10% SDS-polyacrylamide gels show affinity-purified anti–GST IgG and Fab antibodies (left), and affinity-purified anti–Lva IgG and Fab antibodies (right). About 5 μg of each sample was loaded in the presence of β-mercaptoethanol. Arrows indicate intact heavy chains (HC) and light chains (LC), in addition to heavy chain and light chain fragments, HC Fab and LC Fab, respectively. (Bottom) Two Myo-GFP embryos shown 65 min after being injected at the beginning of nuclear cycle 14 with either anti–GST Fab (5 mg/ml, left), or anti–Lva Fab (8.5 mg/ml, right) antibodies. The large Myo-GFP puncta observed in anti–Lva IgG-injected embryos are not observed in anti–Lva Fab antibody-injected embryos. The white arrowheads indicate the injection site. Views are sagittal. Bar, 10 μm.
Figure 8
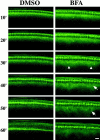
BFA inhibits furrow progression. (Left) DMSO-injected embryo undergoing normal furrowing. (Right) BFA-injected embryo displaying arrested furrowing at the fast phase. Arrowheads indicate unusually high levels of subcortical Myo-GFP, and arrows indicate a defective nucleus. Time (minutes) is relative to the start of cycle 14. The dorsal, injected surface of each embryo is shown. All views are sagittal. Bar, 10 μm.
Comment in
- The ups and downs of life in an epithelium.
Krämer H. Krämer H. J Cell Biol. 2000 Nov 13;151(4):F15-8. doi: 10.1083/jcb.151.4.f15. J Cell Biol. 2000. PMID: 11076958 Free PMC article. Review. No abstract available.
Similar articles
- The golgin Lava lamp mediates dynein-based Golgi movements during Drosophila cellularization.
Papoulas O, Hays TS, Sisson JC. Papoulas O, et al. Nat Cell Biol. 2005 Jun;7(6):612-8. doi: 10.1038/ncb1264. Epub 2004 May 22. Nat Cell Biol. 2005. PMID: 15908943 - Evidence for functional differentiation among Drosophila septins in cytokinesis and cellularization.
Adam JC, Pringle JR, Peifer M. Adam JC, et al. Mol Biol Cell. 2000 Sep;11(9):3123-35. doi: 10.1091/mbc.11.9.3123. Mol Biol Cell. 2000. PMID: 10982405 Free PMC article. - Polarized insertion of new membrane from a cytoplasmic reservoir during cleavage of the Drosophila embryo.
Lecuit T, Wieschaus E. Lecuit T, et al. J Cell Biol. 2000 Aug 21;150(4):849-60. doi: 10.1083/jcb.150.4.849. J Cell Biol. 2000. PMID: 10953008 Free PMC article. - Membrane-actin interactions in morphogenesis: Lessons learned from Drosophila cellularization.
Sokac AM, Biel N, De Renzis S. Sokac AM, et al. Semin Cell Dev Biol. 2023 Jan 15;133:107-122. doi: 10.1016/j.semcdb.2022.03.028. Epub 2022 Apr 5. Semin Cell Dev Biol. 2023. PMID: 35396167 Free PMC article. Review. - A spectrin membrane skeleton of the Golgi complex.
Beck KA, Nelson WJ. Beck KA, et al. Biochim Biophys Acta. 1998 Aug 14;1404(1-2):153-60. doi: 10.1016/s0167-4889(98)00054-8. Biochim Biophys Acta. 1998. PMID: 9714784 Review.
Cited by
- Jagunal is required for reorganizing the endoplasmic reticulum during Drosophila oogenesis.
Lee S, Cooley L. Lee S, et al. J Cell Biol. 2007 Mar 26;176(7):941-52. doi: 10.1083/jcb.200701048. J Cell Biol. 2007. PMID: 17389229 Free PMC article. - The organization of Golgi in Drosophila bristles requires microtubule motor protein function and a properly organized microtubule array.
Melkov A, Baskar R, Shachal R, Alcalay Y, Abdu U. Melkov A, et al. PLoS One. 2019 Oct 2;14(10):e0223174. doi: 10.1371/journal.pone.0223174. eCollection 2019. PLoS One. 2019. PMID: 31577833 Free PMC article. - The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1(+)-encoding syntaxin-like protein.
Nakamura T, Nakamura-Kubo M, Hirata A, Shimoda C. Nakamura T, et al. Mol Biol Cell. 2001 Dec;12(12):3955-72. doi: 10.1091/mbc.12.12.3955. Mol Biol Cell. 2001. PMID: 11739793 Free PMC article. - Here come the septins: novel polymers that coordinate intracellular functions and organization.
Spiliotis ET, Nelson WJ. Spiliotis ET, et al. J Cell Sci. 2006 Jan 1;119(Pt 1):4-10. doi: 10.1242/jcs.02746. J Cell Sci. 2006. PMID: 16371649 Free PMC article. Review. - GOLPH3 is essential for contractile ring formation and Rab11 localization to the cleavage site during cytokinesis in Drosophila melanogaster.
Sechi S, Colotti G, Belloni G, Mattei V, Frappaolo A, Raffa GD, Fuller MT, Giansanti MG. Sechi S, et al. PLoS Genet. 2014 May 1;10(5):e1004305. doi: 10.1371/journal.pgen.1004305. eCollection 2014 May. PLoS Genet. 2014. PMID: 24786584 Free PMC article.
References
- Afshar K., Stuart B., Wasserman S.A. Functional analysis of the Drosophila Diaphanous FH protein in early embryonic development. Development (Camb.) 2000;127:1887–1897. - PubMed
- Beck K.A., Nelson W.J. A spectrin membrane skeleton of the Golgi complex. Biochim. Biophys. Acta. 1998;1404:153–160. - PubMed
- Beites C.L., Xie H., Bowser R., Trimble W.S. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 1999;2:434–439. - PubMed
- Bhat M.A., Izaddoost S., Lu Y., Cho K.O., Choi K.W., Bellen H.J. Discs Lost, a novel multi-PDZ domain protein, establishes and maintains epithelial polarity. Cell. 1999;96:833–845. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM019200/GM/NIGMS NIH HHS/United States
- GM46409-08/GM/NIGMS NIH HHS/United States
- R01 GM023928/GM/NIGMS NIH HHS/United States
- GM23928/GM/NIGMS NIH HHS/United States
- R01 GM046409/GM/NIGMS NIH HHS/United States
- 1F32GM19200-01/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous