The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes - PubMed (original) (raw)
The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes
Y Xue et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The SWI/SNF family of chromatin-remodeling complexes facilitates gene expression by helping transcription factors gain access to their targets in chromatin. SWI/SNF and Rsc are distinctive members of this family from yeast. They have similar protein components and catalytic activities but differ in biological function. Rsc is required for cell cycle progression through mitosis, whereas SWI/SNF is not. Human complexes of this family have also been identified, which have often been considered related to yeast SWI/SNF. However, all human subunits identified to date are equally similar to components of both SWI/SNF and Rsc, leaving open the possibility that some or all of the human complexes are rather related to Rsc. Here, we present evidence that the previously identified human SWI/SNF-B complex is indeed of the Rsc type. It contains six components conserved in both Rsc and SWI/SNF. Importantly, it has a unique subunit, BAF180, that harbors a distinctive set of structural motifs characteristic of three components of Rsc. Of the two mammalian ATPases known to be related to those in the yeast complexes, human SWI/SNF-B contains only the homolog that functions like Rsc during cell growth. Immunofluorescence studies with a BAF180 antibody revealed that SWI/SNF-B localizes at the kinetochores of chromosomes during mitosis. Our data suggest that SWI/SNF-B and Rsc represent a novel subfamily of chromatin-remodeling complexes conserved from yeast to human, and could participate in cell division at kinetochores of mitotic chromosomes.
Figures
Figure 1
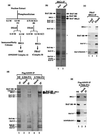
BAF180 distinguishes two similar human chromatin-remodeling complexes. (a) A diagram of the purification scheme for BAF and PBAF complexes (20). Previously assigned names for these complexes are given in parentheses. (b) A silver-stained SDS-gel showing BAF and PBAF purified with BRG1 antibody. Unique components are marked with arrows. The band designated as BAF240 (open arrow) was found to be present in substoichiometric amounts by both BRG1 and BAF180 antibodies (see Fig. 3_a_). Some IgG and crosslinked-IgG are indicated with an asterisk. (c) Immunoblot analysis of the load, flowthrough (FT), and eluate fractions from the BRG1 antibody column. The doublet bands of BAF180 may represent gene products derived from alternatively spliced mRNA. (d) A silver-stained SDS-gel of the human SWI/SNF complex A (lane 3) and B (lane 5) isolated by using a monoclonal antibody against the flag-tagged hSNF5 (34). Immunopurification was performed by using either unfractionated nuclear extract (NE), or fractions from phophocellulose (P11) fractionation of the nuclear extract as indicated on top of the figure (lanes 2–5). Mock purifications (lanes 2 and 4) were done by using extracts derived from regular HeLa cells that do not express the flag-hSNF5. BAF200 may correspond to the protein referred to as p210 in ref. . A polypeptide in complex A, marked as “unknown,” appears to be present in the mock purification and could be a contaminant. (e) Immunoblot analysis of the SWI/SNF complex B from Fig. 1_d_. Nuclear extract (lane 1) was shown as a control.
Figure 2
BAF180 is a human homolog of chicken Polybromo and contains structural motifs similar to yeast Rsc1/Rsc2/Rsc4. (a) Schematic representation of domain structures of human BAF180, chicken Polybromo, ORFs from Drosophila (CG11375) and C. elegans (CE05310), and yeast Rsc1/Rsc2/Rsc4. The BAH1 region of BAF180 is about 20 amino acids shorter than the motifs in other proteins. (b and c) Alignment of bromodomains (Br) and BAH regions for hBAF180, chicken PB1 (gBAH), and the three yeast Rsc subunits. A consensus sequence is shown at the bottom. The BAH regions of Ash1 (a Drosophila trithorax group protein) and mta1 (a subunit of nucleosome remodeling and histone deacetylation chromatin-remodeling complex; ref. 33) are shown for comparison. (d) Northern blotting of total RNA from multiple human tissues by using BAF180 or actin DNA as probes.
Figure 3
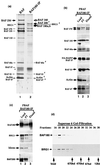
The PBAF complex contains both common and distinctive subunits compared with BAF. (a) A silver-stained SDS-gel showing PBAF purified with BAF180 antibody (lane 2). BAF complex was shown as a control (lane 1). Unique components are marked with arrows. The presence of IgG is indicated by an asterisk. Several polypeptides that have been further identified by mass spectrometry analysis were denoted with #. (b and c) Immunoblot analysis of the load, flowthrough (FT), and eluate fractions from a BAF180 antibody column. The doublet bands of BAF180 may represent isoforms from alternatively spliced mRNA. (d) Immunoblot analysis of elution profile of a Superose 6 gel-filtration chromatography to estimate the molecular mass of PBAF.
Figure 4
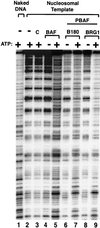
Human PBAF contains an ATP-dependent mononucleosome disruption activity similar to BAF. An autoradiograph from mononucleosome disruption assay with PBAF purified with either BAF180 (B180) or BRG1 antibodies. The result for BAF is shown for comparison. The complexes and templates used are shown above figure. C, A control using antibody beads without complex loaded. The presence (+) or absence (−) of 1 mM ATP is indicated.
Figure 5
PBAF localizes at mitotic kinetochores and spindle poles. Immunofluorescence micrographs of cells in interphase (Top), prometaphase without nocodazole (Middle), and prometaphase in 10 μM nocodazole (Bottom). The small inserts in the lower right-hand corners of the bottom row illustrate colocalization of PBAF and dynein at an enlarged kinetochore. The kinetochore that appears in the zoomed_Inset_ is marked by a box in the image obtained from differential interference contrast light microscopy (DIC). DNA was visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining. In the overlay column, green represents PBAF staining and red represents dynein. The arrows indicate the spindle poles. Scale bar = 5 μm.
Similar articles
- A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex.
Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. Nie Z, et al. Mol Cell Biol. 2000 Dec;20(23):8879-88. doi: 10.1128/MCB.20.23.8879-8888.2000. Mol Cell Biol. 2000. PMID: 11073988 Free PMC article. - RSC, an essential, abundant chromatin-remodeling complex.
Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. Cairns BR, et al. Cell. 1996 Dec 27;87(7):1249-60. doi: 10.1016/s0092-8674(00)81820-6. Cell. 1996. PMID: 8980231 - Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF.
Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Cairns BR, et al. Mol Cell. 1998 Nov;2(5):639-51. doi: 10.1016/s1097-2765(00)80162-8. Mol Cell. 1998. PMID: 9844636 - Promoter targeting and chromatin remodeling by the SWI/SNF complex.
Peterson CL, Workman JL. Peterson CL, et al. Curr Opin Genet Dev. 2000 Apr;10(2):187-92. doi: 10.1016/s0959-437x(00)00068-x. Curr Opin Genet Dev. 2000. PMID: 10753786 Review. - ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job.
Muchardt C, Yaniv M. Muchardt C, et al. J Mol Biol. 1999 Oct 22;293(2):187-98. doi: 10.1006/jmbi.1999.2999. J Mol Biol. 1999. PMID: 10529347 Review.
Cited by
- Chromatin and the genome integrity network.
Papamichos-Chronakis M, Peterson CL. Papamichos-Chronakis M, et al. Nat Rev Genet. 2013 Jan;14(1):62-75. doi: 10.1038/nrg3345. Nat Rev Genet. 2013. PMID: 23247436 Free PMC article. Review. - A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome.
Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. Meetei AR, et al. Mol Cell Biol. 2003 May;23(10):3417-26. doi: 10.1128/MCB.23.10.3417-3426.2003. Mol Cell Biol. 2003. PMID: 12724401 Free PMC article. - Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1alpha.
Nielsen AL, Sanchez C, Ichinose H, Cerviño M, Lerouge T, Chambon P, Losson R. Nielsen AL, et al. EMBO J. 2002 Nov 1;21(21):5797-806. doi: 10.1093/emboj/cdf560. EMBO J. 2002. PMID: 12411497 Free PMC article. - Neuron-specific chromatin remodeling: a missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders.
Vogel-Ciernia A, Wood MA. Vogel-Ciernia A, et al. Neuropharmacology. 2014 May;80:18-27. doi: 10.1016/j.neuropharm.2013.10.002. Epub 2013 Oct 15. Neuropharmacology. 2014. PMID: 24140580 Free PMC article. Review. - Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches.
Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, Rozowsky J, Bhardwaj N, Gerstein MB, Snyder M. Euskirchen GM, et al. PLoS Genet. 2011 Mar;7(3):e1002008. doi: 10.1371/journal.pgen.1002008. Epub 2011 Mar 3. PLoS Genet. 2011. PMID: 21408204 Free PMC article.
References
- Pazin M J, Kadonaga J T. Cell. 1997;88:737–740. - PubMed
- Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:545–579. - PubMed
- Kornberg R D, Lorch Y. Cell. 1999;98:285–294. - PubMed
- Cote J, Quinn J, Workman J L, Peterson C L. Science. 1994;265:53–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous