Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3'-processing apparatus - PubMed (original) (raw)
Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3'-processing apparatus
A Fatica et al. EMBO J. 2000.
Abstract
In Saccharomyces cerevisiae, snoRNAs are encoded by independent genes and within introns. Despite this heterogenous organization, snoRNA biosynthesis relies on a common theme: entry sites for 5'-3' and 3'-5' exonucleases are created on precursor molecules allowing the release of mature snoRNAs. In independently transcribed snoRNAs, such entry sites are often produced by the Rnt1p endonuclease. In many cases, cleavage sites are absent in the 3' portion of the pre-snoRNAs, suggesting that processing starts from the 3' end of the primary transcript. Here we show that cleavage/polyadenylation sites driving efficient polyadenylation, such as CYC1, prevent production of mature and functional snoRNPs. With these sites, snoRNA accumulation is restored only if polyadenylation activity is inhibited. Analysis of sequences downstream of snoRNA-coding units and the use of strains carrying mutations in RNA polymerase II (polII) cleavage/polyadenylation activities allowed us to establish that formation of snoRNA mature 3' ends requires only the cleavage activity of the polII 3'-processing machinery. These data indicate that, in vivo, uncoupling of cleavage and polyadenylation is necessary for an essential cellular biosynthesis.
Figures
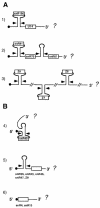
Fig. 1. Schematic representation of the different prototypes of independently transcribed snoRNA-coding units. The boxes represent the snoRNA-coding regions, the stems and arrows indicate the Rnt1p substrates and cleavage sites, respectively, and the dots represent the cap structure. Poly- and monocistronic units are schematically represented in (A) and (B), respectively. In all cases studied so far, the 5′ cap is either present in the mature snoRNA (6) or it is removed by Rnt1p cleavage (1–5) (Chanfreau et al., 1998).
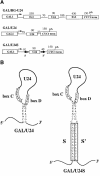
Fig. 2. Schematic representation of the GAL/CYC1 constructs containing the U24 snoRNA-coding sequences. (A) The different boxes indicate the promoter (GAL1), the cleavage/polyadenylation site (CYC1 term), the BEL1 exons (Ex1 and Ex2) and the snoRNA-coding region (U24). Numbers indicate the size of the different portions of the transcripts, and the divergent arrows (S and S′) represent the inverted repeats forming a 13 nucleotide external stem. GAL/HG-U24 contains the U24/EST genomic coding region (Qu et al., 1995) cloned between the GAL and CYC1 sequences of plasmid p413GAL1 (Mumberg et al., 1994). GAL/U24 contains the U24 coding region flanked by 79 and 27 nucleotides of the 5′-untranslated region of GAL1 and of vector sequences, respectively; GAL/U24S contains, in addition, the S/S′ inverted repeats flanking the intron-encoded U18 snoRNA (Villa et al., 2000). (B) Schematic representation of the pre-snoRNAs produced by the GAL/U24 and GAL/U24S constructs. The conserved C and D boxes are indicated together with the terminal stem; the nucleotides present in the mature snoRNA are represented in bold. The external stem is represented by the S and S′ boxes.
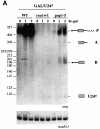
Fig. 3. Processing of the different U24 constructs. (A) Northern blot analysis of 5 µg of total RNA extracted from exponentially grown cells of strain YAF2 (see Materials and methods) either untransformed (lane ΔU24) or transformed with GAL/U24HG (lane GAL/U24HG), GAL/U24 (lane GAL/U24) or GAL/U24S (lane GAL/U24S) plasmids. RNA from strain CH1462 was also loaded as control (lane WT). The probe was a 32P-labeled anti-U24 oligonucleotide. The different products of the reaction and the molecular weight markers (_Msp_I-digested pBR322) are indicated at the sides. The lower panel shows a control hybridization with a U3-specific probe. (B) A 10 µg aliquot of total RNA, extracted from strain YAF2 transformed with GAL/U24 (lane GAL/U24) and GAL/U24S (lane GAL/U24S) plasmids, was incubated in the absence (–) or presence (+) of RNase H and oligo(dT) and analyzed by northern blotting with the U24 probe. (C) Primer extension mapping of the ribose methylations in strain YAF2 either untransformed (lanes ΔU24) or transformed with GAL/U24HG (lanes GAL/U24HG), GAL/U24 (lanes GAL/U24) or GAL/U24S (lanes GAL/U24S) plasmids. A 32P-labeled primer, complementary to nucleotides 1458–1477 of yeast 25S rRNA, was annealed to each RNA sample and extended with AMV reverse transcriptase in the presence of decreasing concentrations of dNTPs (Maden et al., 1995; Kiss-László et al., 1996). The arrows indicate the positions of the modified nucleotides. (D) Tagged (*) versions of the different U24 constructs were utilized in this experiment. Strain YAF2, containing GAL/U24* or GAL/U24*S plasmids, was induced for 2 h in galactose, cells were lysed and immunoprecipitated with IgG beads. RNA was extracted from pellets (P) and supernatants (S) and run on a 6% polyacrylamide–urea gel in parallel with RNA extracted from non-immunoprecipitated extracts (T). Northern blot analysis was performed with a 32P-labeled tag-specific oligonucleotide. The different products of the reaction are indicated at the side by letters, as in (A). The band indicated by a dot represents a degradation product. The lower panels show control hybridizations with U3 snoRNA- and U5 snRNA-specific probes.
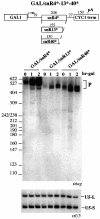
Fig. 4. Analysis of U24 processing in different mutant strains. After 1 h shift at the non-permissive temperature (time 0), cultures were induced with galactose for the indicated times. Northern blot analysis of RNA was performed with the CH1462 strain (WT) or the strains carrying the mutant alleles rna14-1, pap1-5, rnt1Δ, rat1-1 and nup145N transformed with GAL/U24* (A) or GAL/U24*S (B) plasmids. Both blots were hybridized with a 32P-labeled tag-specific oligonucleotide. Size markers (_Msp_I-digested pBR322) and schematic representations of the different processing products are indicated at the sides. The lower panels show control hybridizations with an snR13 snoRNA-specific probe.
Fig. 4. Analysis of U24 processing in different mutant strains. After 1 h shift at the non-permissive temperature (time 0), cultures were induced with galactose for the indicated times. Northern blot analysis of RNA was performed with the CH1462 strain (WT) or the strains carrying the mutant alleles rna14-1, pap1-5, rnt1Δ, rat1-1 and nup145N transformed with GAL/U24* (A) or GAL/U24*S (B) plasmids. Both blots were hybridized with a 32P-labeled tag-specific oligonucleotide. Size markers (_Msp_I-digested pBR322) and schematic representations of the different processing products are indicated at the sides. The lower panels show control hybridizations with an snR13 snoRNA-specific probe.
Fig. 5. Processing activity of monocistronic snoRNAs under a polyadenylation-dependent site. The schematic representation of the constructs utilized is indicated above. The boxes indicate the GAL1 promoter, the CYC1 cleavage/polyadenylation site and the sequences coding for the mature snoRNAs, snR4, snR13 and snR40. The snoRNA-coding regions are flanked by 79 and 27 nucleotides of the 5′-untranslated region of GAL1 and of vector sequences, respectively. All three snoRNAs possess a six or seven nucleotide terminal stem. All these constructs contain the same tag sequence (*) in the snoRNA-coding region. Middle panel: northern blot analysis of RNA, extracted at different times after galactose induction (indicated in hours above the lanes), from the CH1462 strain transformed with GAL/snR4* (lanes GAL/snR4*), GAL/snR13* (lanes GAL/snR13*) and GAL/snR40* (lanes GAL/snR40*) plasmids. The blot was hybridized with a 32P-labeled tag-specific oligonucleotide. Size markers (_Msp_I-digested pBR322) are indicated at the side. The lower panel shows a control hybridization with a U5 snRNA-specific probe.
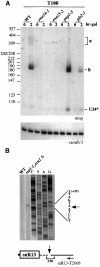
Fig. 6. Characterization of 3′ downstream regions of independently transcribed snoRNAs. (A) Schematic representation of the constructs utilized. The constructs are the same as those shown in Figure 2 up to the CYC region; in the T-type constructs (T200, T100, T50 and T30), the CYC1 cleavage/polyadenylation site was substituted with 200, 100, 50 and 30 nucleotides present in the 3′-flanking region of the snR13 coding sequence (shaded box). Construct 5′stem-T100 contains an Rnt1p-processing site upstream of the U24 coding sequence. The S-T200 plasmid contain the S/S′ external stem as in construct GAL/U24*S. The T0 construct has no snR13 terminator region. Vector sequences are represented by a thick black line. (B and C) Northern blot analysis of RNA, extracted at different times after galactose induction from the CH1462 strain transformed with the plasmids indicated above the lanes. Molecules indicated by the asterisk are polyadenylated products. The lower panels show control hybridizations with an snR13-specific probe. At the bottom of the figure, the schematic representation of the products obtained with the different constructs is indicated. Dotted lines indicate trimming from the available free ends. Rnt1p cleavage sites are indicated by arrows, and pacmen represent 5′–3′ and 3′–5′ exonucleases. Removal of the cap structure, followed by Rnt1p digestion, also occurs but with a very low efficiency (not indicated in the figure).
Fig. 6. Characterization of 3′ downstream regions of independently transcribed snoRNAs. (A) Schematic representation of the constructs utilized. The constructs are the same as those shown in Figure 2 up to the CYC region; in the T-type constructs (T200, T100, T50 and T30), the CYC1 cleavage/polyadenylation site was substituted with 200, 100, 50 and 30 nucleotides present in the 3′-flanking region of the snR13 coding sequence (shaded box). Construct 5′stem-T100 contains an Rnt1p-processing site upstream of the U24 coding sequence. The S-T200 plasmid contain the S/S′ external stem as in construct GAL/U24*S. The T0 construct has no snR13 terminator region. Vector sequences are represented by a thick black line. (B and C) Northern blot analysis of RNA, extracted at different times after galactose induction from the CH1462 strain transformed with the plasmids indicated above the lanes. Molecules indicated by the asterisk are polyadenylated products. The lower panels show control hybridizations with an snR13-specific probe. At the bottom of the figure, the schematic representation of the products obtained with the different constructs is indicated. Dotted lines indicate trimming from the available free ends. Rnt1p cleavage sites are indicated by arrows, and pacmen represent 5′–3′ and 3′–5′ exonucleases. Removal of the cap structure, followed by Rnt1p digestion, also occurs but with a very low efficiency (not indicated in the figure).
Fig. 7. (A) Plasmids T100 and T0 were transformed into strain W303-1B (WT) and into strains carrying the rna14-1, rna15-2, pap1-5 and pfs2-1 mutant alleles (corresponding lanes); after 1 h shift at the non-permissive temperature (time 0), the cultures were induced with galactose for 2 h (time 2 h). RNA was extracted and run on a 6% polyacrylamide gel. Blots were hybridized with a 32P-labeled tag-specific oligonucleotide. Size markers (_Msp_I-digested pBR322) and the different processing products are indicated at the sides. The lower panel shows a control hybridization with an snR13-specific probe. (B) A 10 µg aliquot of total RNA, extracted from the CH1462 strain (lane WT) and from the rat1-1 xrn1Δ double mutant strain (lane rat1-1 xrn1Δ) grown for 2 h at the restrictive temperature, was elongated with reverse transcriptase in the presence of the R13ter200-b primer, which is complementary to a region 200 nucleotides downstream of the snR13 coding region. The sequence around the stop site is shown at the side, while the diagrammatic representation of the cut-off products is shown below together with an indication of the region contained in the T30 construct.
Fig. 8. Analysis of the snR47 terminator. The schematic representation of construct GAL/U24*-47T is shown above. Plasmid was transformed into strain CH1462 (lanes GAL/U24*-47T); after 1 h shift at the non-permissive temperature (time 0), the cultures were induced with galactose for 1 and 2 h. RNA was extracted and run on a 6% polyacrylamide gel in parallel with RNA extracted from cells transformed with GAL/U24* (lanes GAL/U24*). Blots were hybridized with a 32P-labeled tag-specific oligonucleotide. _Msp_I-digested pBR322 and the different processing products are indicated at the sides. The lower panel shows a control hybridization with an snR13-specific probe.
Similar articles
- Pti1p and Ref2p found in association with the mRNA 3' end formation complex direct snoRNA maturation.
Dheur S, Vo le TA, Voisinet-Hakil F, Minet M, Schmitter JM, Lacroute F, Wyers F, Minvielle-Sebastia L. Dheur S, et al. EMBO J. 2003 Jun 2;22(11):2831-40. doi: 10.1093/emboj/cdg253. EMBO J. 2003. PMID: 12773397 Free PMC article. - The roles of endonucleolytic cleavage and exonucleolytic digestion in the 5'-end processing of S. cerevisiae box C/D snoRNAs.
Lee CY, Lee A, Chanfreau G. Lee CY, et al. RNA. 2003 Nov;9(11):1362-70. doi: 10.1261/rna.5126203. RNA. 2003. PMID: 14561886 Free PMC article. - The position of yeast snoRNA-coding regions within host introns is essential for their biosynthesis and for efficient splicing of the host pre-mRNA.
Vincenti S, De Chiara V, Bozzoni I, Presutti C. Vincenti S, et al. RNA. 2007 Jan;13(1):138-50. doi: 10.1261/rna.251907. Epub 2006 Nov 29. RNA. 2007. PMID: 17135484 Free PMC article. - A history of poly A sequences: from formation to factors to function.
Edmonds M. Edmonds M. Prog Nucleic Acid Res Mol Biol. 2002;71:285-389. doi: 10.1016/s0079-6603(02)71046-5. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12102557 Review. - Small nucleolar RNAs.
Eliceiri GL. Eliceiri GL. Cell Mol Life Sci. 1999 Oct 1;56(1-2):22-31. doi: 10.1007/s000180050003. Cell Mol Life Sci. 1999. PMID: 11213258 Free PMC article. Review.
Cited by
- Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast.
Mischo HE, Proudfoot NJ. Mischo HE, et al. Biochim Biophys Acta. 2013 Jan;1829(1):174-85. doi: 10.1016/j.bbagrm.2012.10.003. Epub 2012 Oct 17. Biochim Biophys Acta. 2013. PMID: 23085255 Free PMC article. Review. - Contribution of domain structure to the RNA 3' end processing and degradation functions of the nuclear exosome subunit Rrp6p.
Phillips S, Butler JS. Phillips S, et al. RNA. 2003 Sep;9(9):1098-107. doi: 10.1261/rna.5560903. RNA. 2003. PMID: 12923258 Free PMC article. - A novel plasmid-based microarray screen identifies suppressors of rrp6Delta in Saccharomyces cerevisiae.
Abruzzi K, Denome S, Olsen JR, Assenholt J, Haaning LL, Jensen TH, Rosbash M. Abruzzi K, et al. Mol Cell Biol. 2007 Feb;27(3):1044-55. doi: 10.1128/MCB.01299-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101774 Free PMC article. - Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs.
Dez C, Noaillac-Depeyre J, Caizergues-Ferrer M, Henry Y. Dez C, et al. Mol Cell Biol. 2002 Oct;22(20):7053-65. doi: 10.1128/MCB.22.20.7053-7065.2002. Mol Cell Biol. 2002. PMID: 12242285 Free PMC article. - XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast.
van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Né P, Loeillet S, Nicolas A, Thermes C, Morillon A. van Dijk EL, et al. Nature. 2011 Jun 22;475(7354):114-7. doi: 10.1038/nature10118. Nature. 2011. PMID: 21697827
References
- Amberg D.C., Goldstein,A.L. and Cole,C.N. (1992) Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev., 6, 1173–1189. - PubMed
- Bachellerie J.-P., Michot,B., Nicoloso,M., Balakin,A., Ni,J. and Fournier,M.J. (1995) Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci., 20, 261–264. - PubMed
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous