Mutant prevention concentration as a measure of fluoroquinolone potency against mycobacteria - PubMed (original) (raw)
Mutant prevention concentration as a measure of fluoroquinolone potency against mycobacteria
G Sindelar et al. Antimicrob Agents Chemother. 2000 Dec.
Abstract
Mutant prevention concentration (MPC) has been proposed as a new measure of antibiotic potency by which the ability to restrict selection of resistant mutants is evaluated. To determine whether MPC provides potency information unavailable from the more customary measurement of the MIC, 18 fluoroquinolones were examined for their ability to block the growth of Mycobacterium smegmatis and to select resistant mutants from wild-type populations. Both MPC and MIC were affected by changes in the moiety at the fluoroquinolone C-8 position and in alkyl groups attached to the C-7 piperazinyl ring. When eight resistant mutants, altered in the gyrase A protein, were tested with fluoroquinolones having either a methoxy or a hydrogen at the C-8 position, the MIC for the most resistant mutant correlated better with the MPC than did the MIC for wild-type cells. For C-8-fluorine derivatives, which were generally less active than the C-8-methoxy compounds but which were more active than C-8-hydrogen derivatives, the MICs for both the mutant and the wild type correlated well with the MPCs. Thus, measurement of the MICs for wild-type cells can reflect the ability of a quinolone to restrict the selection of resistance, but often it does not. With the present series of compounds, the most potent contained a C-8-methoxy and a small group attached to the C-7 ring.
Figures
FIG. 1
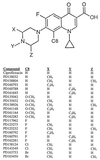
Fluoroquinolone structures. A structure common to the compounds tested is shown, with the variable groups marked X, Y, Z, and C8. Identification numbers for the compounds are listed below the structures. Due to rotation at the C-7 position, substitutions at the 3′ (X) and 5′ (Z) positions are identical.
FIG. 2
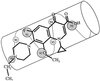
Orientations of fluoroquinolones and GyrA α-helix 4. α-helix 4, adapted from the crystal structure of the breakage-reunion domain of the GyrA protein of E. coli (10), is drawn parallel to the long axis of PD161144. Amino acid numbers represent positions in the M. smegmatis GyrA protein.
FIG. 3
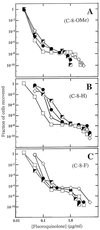
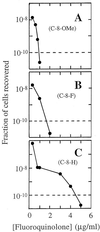
Effect of small C-7-ring alkyls on selection of resistant mutants by fluoroquinolones. M. smegmatis strain KD1163 was applied to agar plates at the indicated fluoroquinolone concentrations. Fluoroquinolones contained a C-8-OMe group (A), a C-8-H group (B), and a C-8-F group (C). Symbols for panel A: squares, PD138926, C-8-OMe C-7-ring dimethyl; diamonds, PD161144, C-8-OMe C-7-ring _N_-ethyl; filled circles, PD161148, C-8-OMe C-7-ring 3′-ethyl; half-filled squares, PD135432, C-8-OMe C-7-ring methyl. Symbols for panel B: squares, PD158804, C-8-H C-7-ring dimethyl; diamonds, PD160788, C-8-H C-7-ring _N_-ethyl; filled circles, PD160793, C-8-H C-7-ring 3′-ethyl; half-filled squares, PD138032, C-8-H C-7-ring methyl. Symbols for panel C, squares, PD125232, C-8-F C-7-ring dimethyl; half-filled squares, PD125275, C-8-F C-7-ring 3′-methyl; diamonds, PD160791, C-8-F C-7-ring _N_-ethyl; filled circles, PD160792, C-8-F C-7-ring 3′-ethyl. A replicate experiment focusing on the second sharp drop in colony recovery gave results similar to those shown.
FIG. 4
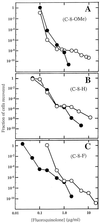
Effect of C-7-ring isopropyl group on selection of resistant mutants by fluoroquinolones. M. smegmatis was applied to agar plates containing the indicated fluoroquinolone concentrations. Fluoroquinolones contained a C-8-OMe group (A), a C-8-H group (B), and a C-8-F group (C). Symbols for panel A: filled circles, PD135042, C-8-OMe with no addition to C-7-ring; open circles, PD162282, C-8-OMe C-7-ring _N_-isopropyl. Symbols for panel B: filled circles, ciprofloxacin, C-8-H with no addition to the C-7 ring; open circles, PD161645, C-8-H C-7-ring _N_-isopropyl. Symbols for panel C: filled circles, PD117962, C-8-F with no addition to C-7 ring; open circles, PD162281, C-8-F C-7-ring _N_-isopropyl. A replicate experiment focusing on the second sharp drop in colony recovery gave results similar to those shown.
FIG. 5
Determination of MPC. Resistant M. smegmatis mutants were recovered from agar plates containing the indicated concentrations of PD161148 (C-8-OMe, C-7-ring 3′-ethyl) (A), PD160792 (C-8-F, C-7-ring 3′-ethyl) (B), and PD160793 (C-8-H, C-7-ring 3′-ethyl) (C). MPC1010 is defined as the concentration at which less than one colony is recovered when 1010 cells are applied to agar plates. MPC1010 is approximated by the intersection of the data curve and the dashed line in the figure.
FIG. 6
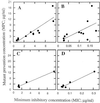
Relationship between MPC and MIC. The MPC of each fluoroquinolone, determined as described in the legend to Fig. 5, was plotted against the MICs of C-8-OMe and C-8-H fluoroquinolones with the C-7-ring moieties indicated in Fig. 2 for the most resistant gyrA mutant (A) or wild-type cells (B). The relationships between the MPC and the MIC for a gyrA mutant or the MIC for the wild type are shown in panels C and D, respectively, for the C-8-F compounds.
Similar articles
- Enhancement of fluoroquinolone activity by C-8 halogen and methoxy moieties: action against a gyrase resistance mutant of Mycobacterium smegmatis and a gyrase-topoisomerase IV double mutant of Staphylococcus aureus.
Lu T, Zhao X, Li X, Drlica-Wagner A, Wang JY, Domagala J, Drlica K. Lu T, et al. Antimicrob Agents Chemother. 2001 Oct;45(10):2703-9. doi: 10.1128/AAC.45.10.2703-2709.2001. Antimicrob Agents Chemother. 2001. PMID: 11557458 Free PMC article. - Engineering the specificity of antibacterial fluoroquinolones: benzenesulfonamide modifications at C-7 of ciprofloxacin change its primary target in Streptococcus pneumoniae from topoisomerase IV to gyrase.
Alovero FL, Pan XS, Morris JE, Manzo RH, Fisher LM. Alovero FL, et al. Antimicrob Agents Chemother. 2000 Feb;44(2):320-5. doi: 10.1128/AAC.44.2.320-325.2000. Antimicrob Agents Chemother. 2000. PMID: 10639357 Free PMC article. - Activities of trovafloxacin compared with those of other fluoroquinolones against purified topoisomerases and gyrA and grlA mutants of Staphylococcus aureus.
Gootz TD, Zaniewski RP, Haskell SL, Kaczmarek FS, Maurice AE. Gootz TD, et al. Antimicrob Agents Chemother. 1999 Aug;43(8):1845-55. doi: 10.1128/AAC.43.8.1845. Antimicrob Agents Chemother. 1999. PMID: 10428901 Free PMC article. - Selection of antibiotic-resistant bacterial mutants: allelic diversity among fluoroquinolone-resistant mutations.
Zhou J, Dong Y, Zhao X, Lee S, Amin A, Ramaswamy S, Domagala J, Musser JM, Drlica K. Zhou J, et al. J Infect Dis. 2000 Aug;182(2):517-25. doi: 10.1086/315708. Epub 2000 Jul 24. J Infect Dis. 2000. PMID: 10915083 - Fluoroquinolones: action and resistance.
Drlica K, Malik M. Drlica K, et al. Curr Top Med Chem. 2003;3(3):249-82. doi: 10.2174/1568026033452537. Curr Top Med Chem. 2003. PMID: 12570763 Review.
Cited by
- In vitro potency and efficacy favor later generation fluoroquinolones for treatment of canine and feline Escherichia coli uropathogens in the United States.
Liu X, Boothe DM, Jin Y, Thungrat K. Liu X, et al. World J Microbiol Biotechnol. 2013 Feb;29(2):347-54. doi: 10.1007/s11274-012-1188-x. Epub 2012 Nov 8. World J Microbiol Biotechnol. 2013. PMID: 23136054 - Fluoroquinolone and quinazolinedione activities against wild-type and gyrase mutant strains of Mycobacterium smegmatis.
Malik M, Marks KR, Mustaev A, Zhao X, Chavda K, Kerns RJ, Drlica K. Malik M, et al. Antimicrob Agents Chemother. 2011 May;55(5):2335-43. doi: 10.1128/AAC.00033-11. Epub 2011 Mar 7. Antimicrob Agents Chemother. 2011. PMID: 21383100 Free PMC article. - Suppression of Emergence of Resistance in Pathogenic Bacteria: Keeping Our Powder Dry, Part 1.
Drusano GL, Louie A, MacGowan A, Hope W. Drusano GL, et al. Antimicrob Agents Chemother. 2015 Dec 28;60(3):1183-93. doi: 10.1128/AAC.02177-15. Antimicrob Agents Chemother. 2015. PMID: 26711759 Free PMC article. Review. - Mutant prevention concentrations of ABT-492, levofloxacin, moxifloxacin, and gatifloxacin against three common respiratory pathogens.
Hermsen ED, Hovde LB, Konstantinides GN, Rotschafer JC. Hermsen ED, et al. Antimicrob Agents Chemother. 2005 Apr;49(4):1633-5. doi: 10.1128/AAC.49.4.1633-1635.2005. Antimicrob Agents Chemother. 2005. PMID: 15793158 Free PMC article. - Synergistic combination of two antimicrobial agents closing each other's mutant selection windows to prevent antimicrobial resistance.
Xu X, Xu L, Yuan G, Wang Y, Qu Y, Zhou M. Xu X, et al. Sci Rep. 2018 May 8;8(1):7237. doi: 10.1038/s41598-018-25714-z. Sci Rep. 2018. PMID: 29740150 Free PMC article.
References
- Bifani P, Plikaytis B B, Kapur V, Stockbauer K, Pan X, Lusfty M, Moghazeh S, Eisner W, Daniel T, Kaplan M, Crawford J T, Musser J M, Kreiswirth B N. Origin and interstate spread of a New York City multidrug resistant Mycobacterium tuberculosis clone family: adverse implications for tuberculosis control in the 21st century. JAMA. 1996;275:452–457. - PubMed
- Bishop O N. Statistics for biology. New York, N.Y: Houghton-Mifflin Co.; 1966. p. 65.
- Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S, Eiglmeier K, Gas S, Barry C E, Tekaia F, Babcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous