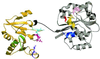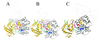Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase - PubMed (original) (raw)
Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase
J M Caruthers et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The eukaryotic translation initiation factor 4A (eIF4A) is a member of the DEA(D/H)-box RNA helicase family, a diverse group of proteins that couples an ATPase activity to RNA binding and unwinding. Previous work has provided the structure of the amino-terminal, ATP-binding domain of eIF4A. Extending those results, we have solved the structure of the carboxyl-terminal domain of eIF4A with data to 1.75 A resolution; it has a parallel alpha-beta topology that superimposes, with minor variations, on the structures and conserved motifs of the equivalent domain in other, distantly related helicases. Using data to 2.8 A resolution and molecular replacement with the refined model of the carboxyl-terminal domain, we have completed the structure of full-length eIF4A; it is a "dumbbell" structure consisting of two compact domains connected by an extended linker. By using the structures of other helicases as a template, compact structures can be modeled for eIF4A that suggest (i) helicase motif IV binds RNA; (ii) Arg-298, which is conserved in the DEA(D/H)-box RNA helicase family but is absent from many other helicases, also binds RNA; and (iii) motifs V and VI "link" the carboxyl-terminal domain to the amino-terminal domain through interactions with ATP and the DEA(D/H) motif, providing a mechanism for coupling ATP binding and hydrolysis with conformational changes that modulate RNA binding.
Figures
Figure 1
Structure of the carboxyl-terminal domain of eIF4A. (A) Stereoview, ribbon drawing of the structure. Conserved motifs are colored as follows: motif IV, VIFCNTRR, residues 263–270, green; “conserved R” motif, residue Arg-298, purple; motif V, RGID, residues 321–324, magenta; motif VI, HRIGRGGR, residues 345–352, cyan. The strands of the β-sheet are labeled sequentially. This and subsequent ribbon drawings were prepared with
molscript
(34) and rendered with
raster
3
d
(35). (B) Topology diagram of the structure. β-Strands are shown as arrows; α-helices, as cylinders. β-Strands and α-helices are labeled sequentially as 1–7 and α1– α6, respectively. Sequences of the conserved motifs are shown in boxes; residues whose side chains are illustrated in_A_ and subsequent figures are underlined.
Figure 2
Ribbon drawing of the structure of full-length eIF4A. The amino- and carboxyl-terminal domains are colored silver and gold, respectively; the 11-residue linker connecting them is colored black. The conserved amino-terminal motifs are colored as follows: motif I, Walker A motif ASQSGTGKT, residues 65–72, blue; motif Ia, PTRELA, residues 97–102, yellow; GG, residues 125–126, orange; TPGR, residues 145–148, pink; motif II, Walker B motif DEAD, residues 169–172, red; motif III, SAT, residues 200–202, green. The conserved carboxyl-terminal motifs are colored as described in Fig. 1_A_.
Figure 3
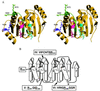
Overlay of amino- and carboxyl-terminal domains of eIF4A onto the equivalent domains of other helicases. The amino-terminal domain of eIF4A is in approximately the same orientation as in Fig. 2. The color convention for the domains and conserved motifs of eIF4A is the same as in Fig. 2. In all cases, the “target” helicase is shown as a transparent tube drawing against the solid eIF4A model. Oligonucleotides are shown as CPK space-filling models. (A) PcrA DNA helicase (PDB ID code 3PJR) with ATP and the single-stranded portion of the DNA. ATP is shown as a ball-and-stick representation. (B) HCV RNA helicase with ssDNA (PDB ID code 1A1V). (C) UvrD DNA helicase (PDB ID code 1D9X).
Similar articles
- Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase.
Johnson ER, McKay DB. Johnson ER, et al. RNA. 1999 Dec;5(12):1526-34. doi: 10.1017/s1355838299991410. RNA. 1999. PMID: 10606264 Free PMC article. - The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis.
Pause A, Méthot N, Sonenberg N. Pause A, et al. Mol Cell Biol. 1993 Nov;13(11):6789-98. doi: 10.1128/mcb.13.11.6789-6798.1993. Mol Cell Biol. 1993. PMID: 8413273 Free PMC article. - The DEAD-box helicase eIF4A: paradigm or the odd one out?
Andreou AZ, Klostermeier D. Andreou AZ, et al. RNA Biol. 2013 Jan;10(1):19-32. doi: 10.4161/rna.21966. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995829 Free PMC article. Review. - eIF4A: the godfather of the DEAD box helicases.
Rogers GW Jr, Komar AA, Merrick WC. Rogers GW Jr, et al. Prog Nucleic Acid Res Mol Biol. 2002;72:307-31. doi: 10.1016/s0079-6603(02)72073-4. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12206455 Review.
Cited by
- Unraveling the 'DEAD-box' helicases of Plasmodium falciparum.
Tuteja R, Pradhan A. Tuteja R, et al. Gene. 2006 Jul 5;376(1):1-12. doi: 10.1016/j.gene.2006.03.007. Epub 2006 Apr 7. Gene. 2006. PMID: 16713133 Free PMC article. Review. - From unwinding to clamping - the DEAD box RNA helicase family.
Linder P, Jankowsky E. Linder P, et al. Nat Rev Mol Cell Biol. 2011 Jul 22;12(8):505-16. doi: 10.1038/nrm3154. Nat Rev Mol Cell Biol. 2011. PMID: 21779027 Review. - Functionally important segments in proteins dissected using Gene Ontology and geometric clustering of peptide fragments.
Manikandan K, Pal D, Ramakumar S, Brener NE, Iyengar SS, Seetharaman G. Manikandan K, et al. Genome Biol. 2008;9(3):R52. doi: 10.1186/gb-2008-9-3-r52. Epub 2008 Mar 10. Genome Biol. 2008. PMID: 18331637 Free PMC article. - Parts, assembly and operation of the RIG-I family of motors.
Rawling DC, Pyle AM. Rawling DC, et al. Curr Opin Struct Biol. 2014 Apr;25:25-33. doi: 10.1016/j.sbi.2013.11.011. Epub 2013 Dec 20. Curr Opin Struct Biol. 2014. PMID: 24878341 Free PMC article. Review. - Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif.
Hardin JW, Hu YX, McKay DB. Hardin JW, et al. J Mol Biol. 2010 Sep 17;402(2):412-27. doi: 10.1016/j.jmb.2010.07.040. Epub 2010 Jul 29. J Mol Biol. 2010. PMID: 20673833 Free PMC article.
References
- Gorbalenya A E, Koonin E V. Curr Opin Struct Biol. 1993;3:419–429.
- Staley J P, Guthrie C. Cell. 1998;92:315–326. - PubMed
- Venema J, Tollervey D. Yeast. 1995;11:1629–1650. - PubMed
- Py B, Higgins C F, Krisch H M, Carpousis A J. Nature (London) 1996;381:169–172. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous