Novel splice variants of cyclin E with altered substrate specificity - PubMed (original) (raw)
Novel splice variants of cyclin E with altered substrate specificity
D C Porter et al. Nucleic Acids Res. 2000.
Abstract
Cyclin E, a G(1) cyclin, is overexpressed and present in low molecular weight (LMW) isoforms in breast cancer cells and tumor tissues. In this study we have examined the possibility that the shortened mRNA splice variants could give rise to tumor-specific cyclin E LMW proteins. We used the Splice Capture method to identify, enumerate and isolate known spliced mRNAs and to look for previously undetected mRNA forms of cyclin E that might be translated into the LMW proteins. We show that a new splice variant of cyclin E found in tumor cells isolated by the Splice Capture strategy, named Delta48, activates CDK2 more robustly than full-length cyclin E when assayed from transiently transfected cells with the natural substrate GST-Rb. We also found the Splice Capture method to be superior to the conventional RNase protection assay in analyzing the cyclin E mRNA present in normal and tumor cells. Splice Capture enumerated the relative abundance of known forms of cyclin E mRNA and easily discovered new splice variants in both normal and tumor cells. We conclude that the abundance of cyclin E splice variants in cells may represent a novel form of regulation of cyclin E, and if translated they show altered substrate specificity compared to the full length form of cyclin E.
Figures
Figure 1
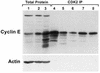
Cyclin E LMW forms bind to and immunoprecipitate with CDK2. Protein was extracted from the 76N normal (lanes 1 and 7), MCF-10A normal (lanes 2 and 8), MDA-MB-157 cancer (lanes 3 and 4), MDA-MB-436 cancer (lane 5) and MCF-7 cancer (lane 6) cell lines. For western blotting 50 µg total protein was loaded (lanes 1–3). Analysis with anti-actin antibody reveals equal loading of total protein. For CDK2 immunoprecipitation (lanes 4–8) 200 µg total cell extract were incubated with a polyclonal antibody to CDK2 cross-linked to protein A–Sepharose. The immune complexes were washed, boiled in reducing sample buffer and western immunoblotted alongside the total cell extracts.
Figure 2
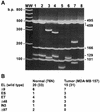
RT–PCR library analysis shows an equal representation of cyclin EL and its splice variants in normal and tumor cells. RT–PCR was performed on normal (76N) and tumor (MDA-MB-157) mRNA using 3′ and 5′ flanking oligonucleotides to the cyclin EL cDNA sequence. The PCR products representing a sample of the cyclin E mRNA species were then cloned into the PCR vector, transformed into E.coli, plated, filter lifted, probed with the 32P-labeled cyclin E probe, positive colonies picked, plasmids prepared, PCR performed again to create a clean product for restriction digest with _Sau_3AI, electrophoresed on 5% acrylamide gels and stained with ethidium bromide. (A) The gel represents the different cyclin E splice variants observed in both normal and tumor cells and the identities were confirmed by sequencing. MW, 100 bp ladder; lane 1, pLXSN-EL plasmid containing the EL form of cyclin E; lane 2, EL, cDNA identical to pLXSN-EL; lane 3, Δ9 + ES, cDNA containing the Δ9 splice variant in combination with the ES splice variant; lane 4, ET, with 135 bp lost from position 706 to 840 (gccctt…gcagag); lane 5, Δ48, with 48 bp lost from position 22 to 69 (cgggat…gcggag); lane 6, Δ9; lane 7, IN3, with 190 bp of genomic sequence from intron 3 which failed to be spliced (bases 1999–2190 of the genomic sequence, accgttGTGAGT…TGTTAGtttttg) (21); lane 8, Δ97, with 97 bp lost from position 24 to 120 (ggatgc…ttgcag). The numbers to the right of the gel identify the length in bp of the _Sau_3AI restriction fragments from the EL cDNA PCR product. (B) The occurrence of each individual variation is shown in normal (76N) and tumor (MDA-MB-157) cells. The number in parentheses indicates the total number of clones examined for each cell line. The ET and Δ48 variants contain an open reading frame and could be translated into a cyclin E protein. IN3 and Δ97 contain a frameshift precluding complete translation from the M1 start. Some cDNAs contained more than one splice variation and these were counted individually.
Figure 3
Expression of cyclin E mRNA in breast cancer cell lines. (A) Schematic representation of the anitsense RNA probes within the cyclin E cDNA used to quantitate cyclin E mRNAs by RNase protection assay in normal (76N and MCF-10A) and tumor (MCF-7, MDA-MB-157, MDA-MB-231, MDA-MB-436 and ZR75T) cells. (B) RNase protection assay with antisense probe 3B in normal and tumor cells. The numbers represent copies of mRNA per cell as quantitated after gel electrophoresis by Cerenkov counts of the protected 32P-labeled antisense RNA probe (± SD). (C) Four overlapping antisense RNA probes were used to quantitate the amount of mRNA and look for the presence of splice variants in MDA-MB-157 cells. (D) Autoradiograph of an RNase protection assay using RNA from a variety of cell lines and the antisense RNA probe 7B. Probe 7B extends the range of protected mRNA (see Fig. 2A) and detects the presence of a smaller protected fragment, possibly due to a low abundance splice variant. This pattern was similar in all cell lines examined.
Figure 4
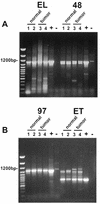
General expression of novel splice variants of cyclin E in normal and tumor breast cells. RT–PCR amplification of the cyclin E coding sequence from normal and tumor-derived breast epithelial cell lines using oligonucleotides UFCE and CE1247 for EL, C3FOR and CE1247 for Δ48, C31FOR and CE1247 for Δ97 and UFCE and ETREV for ET. The cell lines used were as follows: lane 1, MCF-10A; lane 2, 76N; lane 3, MDA-MB-157; lane 4, MDA-MB-436. Normal cells are represented in lanes 1 and 2 and tumor-derived cell lines in lanes 3 and 4. + indicates a positive control using vectors with the appropriate splice variant cDNA inserts as template. – indicates a negative control using the EL vector as template. The negative control for EL was an empty vector. The first lane of each gel shows the 100 bp ladder molecular weight size markers. Note that the only difference observed between normal and tumor cells was the detection of slower migrating ET primed RT–PCR products which were tumor specific and distinguishable from the ET products based on size.
Figure 5
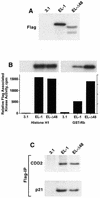
The Δ48 cyclin E splice variant is functionally active. 76N-E6 cells were transiently transfected with cyclin EL–FLAG, Δ48–FLAG or the vector backbone (pCDNA3.1) using the FuGENE reagent. Twenty-four hours following transfection cells were harvested and cell lysates were prepared and subjected to (A) western blot analysis, (B) FLAG kinase analysis and (C) FLAG immune complex formation. For western blot analysis 50 µg protein extract from each condition were analyzed by western blotting with the anti-FLAG antibody. The blot was developed with chemiluminescence reagents. For FLAG kinase activity equal amounts of protein (250 µg) from cell lysates were prepared for each condition and immunoprecipitated with anti-FLAG antibody (polyclonal) coupled to protein A beads, using histone H1 or GST-Rb as substrate. For each condition we show the resulting autoradiogram of the histone H1 and GST-Rb SDS–PAGE and the quantitation of the histone H1 and GST-Rb associated kinase activities by Cerenkov counting. For immunoprecipitation followed by western blot analysis equal amounts of protein (250 µg) from cell lysates prepared for each condition were immunoprecipitated with anti-FLAG (polyclonal) antibody coupled to protein A beads and the immunoprecipitates were subjected to western blot analysis with CDK2 and p21 antibodies.
Similar articles
- Activation of cyclin-dependent kinase 2 by full length and low molecular weight forms of cyclin E in breast cancer cells.
Harwell RM, Mull BB, Porter DC, Keyomarsi K. Harwell RM, et al. J Biol Chem. 2004 Mar 26;279(13):12695-705. doi: 10.1074/jbc.M313407200. Epub 2003 Dec 29. J Biol Chem. 2004. PMID: 14701826 - The low molecular weight (LMW) isoforms of cyclin E deregulate the cell cycle of mammary epithelial cells.
Wingate H, Bedrosian I, Akli S, Keyomarsi K. Wingate H, et al. Cell Cycle. 2003 Sep-Oct;2(5):461-6. Cell Cycle. 2003. PMID: 12963845 - [Detection of cell cycle related gene expression in cultured immortalized human lens epithelial cell].
Wu M, Li S, Zeng J, Gao J, Liu Y. Wu M, et al. Zhonghua Yan Ke Za Zhi. 2002 Jun;38(6):367-71. Zhonghua Yan Ke Za Zhi. 2002. PMID: 12139816 Chinese. - Cyclin E is the only cyclin-dependent kinase 2-associated cyclin that predicts metastasis and survival in early stage non-small cell lung cancer.
Müller-Tidow C, Metzger R, Kügler K, Diederichs S, Idos G, Thomas M, Dockhorn-Dworniczak B, Schneider PM, Koeffler HP, Berdel WE, Serve H. Müller-Tidow C, et al. Cancer Res. 2001 Jan 15;61(2):647-53. Cancer Res. 2001. PMID: 11212263 - Lower molecular weight forms of cyclin E--super activators of the cell cycle?
Stighall M, Berglund P, Landberg G. Stighall M, et al. Cell Cycle. 2003 Sep-Oct;2(5):458-60. Cell Cycle. 2003. PMID: 12963844 Review. No abstract available.
Cited by
- Among B-type cyclins only CLB5 and CLB6 promote premeiotic S phase in Saccharomyces cerevisiae.
DeCesare JM, Stuart DT. DeCesare JM, et al. Genetics. 2012 Mar;190(3):1001-16. doi: 10.1534/genetics.111.134684. Epub 2011 Dec 29. Genetics. 2012. PMID: 22209902 Free PMC article. - Cell cyclins: triggering elements of cancer or not?
Stamatakos M, Palla V, Karaiskos I, Xiromeritis K, Alexiou I, Pateras I, Kontzoglou K. Stamatakos M, et al. World J Surg Oncol. 2010 Dec 22;8:111. doi: 10.1186/1477-7819-8-111. World J Surg Oncol. 2010. PMID: 21176227 Free PMC article. Review. - Targeting cyclin-dependent kinases in human cancers: from small molecules to Peptide inhibitors.
Peyressatre M, Prével C, Pellerano M, Morris MC. Peyressatre M, et al. Cancers (Basel). 2015 Jan 23;7(1):179-237. doi: 10.3390/cancers7010179. Cancers (Basel). 2015. PMID: 25625291 Free PMC article. Review. - An intron-retaining splice variant of human cyclin A2, expressed in adult differentiated tissues, induces a G1/S cell cycle arrest in vitro.
Honda A, Valogne Y, Bou Nader M, Bréchot C, Faivre J. Honda A, et al. PLoS One. 2012;7(6):e39249. doi: 10.1371/journal.pone.0039249. Epub 2012 Jun 20. PLoS One. 2012. PMID: 22745723 Free PMC article. - Genome-wide analysis of core cell cycle genes in Arabidopsis.
Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D. Vandepoele K, et al. Plant Cell. 2002 Apr;14(4):903-16. doi: 10.1105/tpc.010445. Plant Cell. 2002. PMID: 11971144 Free PMC article.
References
- Sheaff R.J. (1997) Methods Enzymol., 283, 173–193. - PubMed
- Pavletich N.P. (1999) J. Mol. Biol., 287, 821–828. - PubMed
- Steeg P.S. and Zhou,Q. (1998) Breast Cancer Res. Treat., 52, 17–28. - PubMed
- Keyomarsi K., Conte,D., Toyofuku,W. and Fox,M.P. (1995) Oncogene, 11, 941–950. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials