A new screen for protein interactions reveals that the Saccharomyces cerevisiae high mobility group proteins Nhp6A/B are involved in the regulation of the GAL1 promoter - PubMed (original) (raw)
A new screen for protein interactions reveals that the Saccharomyces cerevisiae high mobility group proteins Nhp6A/B are involved in the regulation of the GAL1 promoter
H Laser et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The split-ubiquitin assay detects protein interactions in vivo. To identify proteins interacting with Gal4p and Tup1p, two transcriptional regulators, we converted the split-ubiquitin assay into a generally applicable screen for binding partners of specific proteins in vivo. A library of genomic Saccharomyces cerevisiae DNA fragments fused to the N-terminal half of ubiquitin was constructed and transformed into yeast strains carrying either Gal4p or Tup1p as a bait. Both proteins were C-terminally extended by the C-terminal half of ubiquitin followed by a modified Ura3p with an arginine in position 1, a destabilizing residue in the N-end rule pathway. The bait fusion protein alone is stable and enzymatically active. However, upon interaction with its prey, a native-like ubiquitin is reconstituted. RUra3p is then cleaved off by the ubiquitin-specific proteases and rapidly degraded by the N-end rule pathway. In both screens, Nhp6B was identified as a protein in close proximity to Gal4p as well as to Tup1p. Direct interaction between either protein and Nhp6B was confirmed by coprecipitation assays. Genetic analysis revealed that Nhp6B, a member of the HMG1 family of DNA-binding proteins, can influence transcriptional activation as well as repression at a specific locus in the chromosome of the yeast S. cerevisiae.
Figures
Figure 1
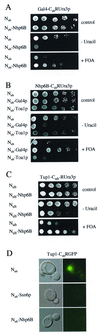
A system to select for protein interactions in vivo. (A) The split-ubiquitin system. Ubiquitin, fused to the N terminus of Ura3p displaying an arginine as its first amino acid (RUra3p), is recognized by the UBPs (line 1). The cleaved RUra3p is rapidly degraded by the N-end rule pathway of protein degradation (line 4). No cleavage of RUra3p takes place if only the C-terminal half of ubiquitin (Cub) is fused between Gal4p and RUra3p (line 2). A protein X is attached to the N-terminal half of ubiquitin. If X interacts with Gal4p, the two coupled Ub peptides are forced into close proximity, a ubiquitin-like molecule is reconstituted, and cleavage by the UBPs is observed (line 3). The freed RUra3p reporter is now rapidly degraded by the enzymes of the N-end rule, resulting in uracil auxotrophy and FOA resistance (line 4). (B) Gal4p interacts with Gal80p in vivo. Shown are serial dilutions of cells coexpressing Nub or a Nub-Gal80p fusion together with Gal4(1–147 + 768–881)-Cub-RUra3p on plates lacking tryptophan and leucine (Top), additionally lacking uracil (Middle), or containing FOA (Bottom). All proteins were expressed from single-copy vectors. (C) Tup1p interacts with Ssn6p in vivo. Shown are serial dilutions of cells coexpressing the depicted Nub and Cub fusions on plates lacking tryptophan and leucine (Upper) or on plates additionally lacking uracil (Lower). All proteins were expressed from single-copy vectors.
Figure 2
Nhp6B was isolated in two independent split-ubiquitin screens using Gal4p or Tup1p as Cub-RUra3 baits. (A) Gal4p interacts with Nhp6B in vivo. Serial dilutions of cells coexpressing Nub or an Nub-Nhp6B fusion together with a fusion of the DNA-binding and activation domains of Gal4(1–147 + 768–881)p to Cub-RUra3p were grown on plates lacking tryptophan and leucine (Top), on plates additionally lacking uracil (Middle), or on plates containing FOA (Bottom). Nub and Nub fused to full-length Nhp6B were expressed from multicopy vectors. (B) The activation domain of Gal4p is sufficient for the interaction with Nhp6B. Serial dilutions of cells coexpressing Nub, Nub fused to the activation domain of Gal4p (amino acids 768–881; Nub-Gal4p), or Nub attached to the large subunit of TFIIA (Nub-Toa1p) together with Nhp6B-Cub-RUra3p were grown on plates lacking tryptophan and leucine (Top), on plates additionally lacking uracil (Middle), or on plates containing FOA (Bottom). Nub, Nub-Gal4p, and Nub-Toa1p were expressed from multicopy vectors. (C) Tup1p interacts with Nhp6B in vivo. Serial dilutions of cells coexpressing the depicted Nub and Cub fusions were grown on plates lacking tryptophan and leucine (Top), on plates additionally lacking uracil (Middle), or on plates containing FOA (Bottom). Nub and the clone isolated from the library expressing Nub-Nhp6B that lacked the first 22 amino acids of Nhp6B were on multicopy vectors. (D) Tup1-Cub-RGFP is located in the nucleus and interacts with Nub-Ssn6p and Nub-Nhp6B. Cells expressing the depicted fusions from single-copy vectors were analyzed under a Leitz fluorescence microscope with phase contrast (Left) and fluorescence (Right).
Figure 3
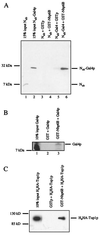
Nhp6B interacts with Gal4p and Tup1p in vitro. (A) Gal4p coprecipitates together with Nhp6B from_S. cerevisiae_ extracts. Extracts from S. cerevisiae cells expressing Nub or Nub-Gal4p (amino acids 768–881) from multicopy vectors were incubated with GSTp or GST-Nhp6B purified from E. coli on glutathione beads. Coprecipitated proteins were separated on an SDS gel and visualized on a Western blot with an anti-HA antibody with the help of an HA tag present in the Nub moiety. (B) In vitro translated Gal4p interacts with Nhp6B. The activation domain of Gal4p (amino acids 768–881) was radiolabeled by in vitro translation and incubated with a bacterially purified GSTp or a GST-Nhp6B fusion bound to glutathione beads. Coprecipitated proteins were visualized by autoradiography. A truncated form of the activation domain of Gal4p, migrating faster in the SDS gel, showed no interaction with GST-Nhp6B. (C) Purified Tup1p interacts with purified Nhp6B. A H6HA-Tup1p fusion was purified on an Ni column and incubated with purified GSTp or GST-Nhp6B on glutathione beads. Coprecipitated H6HA-Tup1p was visualized on a Western blot with an anti-HA antibody.
Figure 4
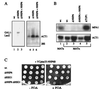
The interaction between Nhp6B and Tup1p is biologically relevant. (A) Nhp6 is necessary for glucose repression of the GAL1 promoter. RNA was prepared from the depicted strains carrying a GAL1-LacZ fusion integrated at the GAL1 locus. JD53 was used as wild-type parental strain (lanes 1 and 4). The ΔNHP6 strain was derived from JD53 that lacks NHP6A and NHP6B (lanes 2 and 5). In the strain ΔNHP6 + NHP6 (lanes 3 and 6), NHP6A and NHP6B had been reintegrated into the original loci. Equal amounts of total RNA were loaded as confirmed by ethidium bromide staining (not shown) and background hybridization to the 28 S rRNA (Right). The Northern blot was probed with a LacZ probe (lanes 1–3) and with an ACT1 probe (lanes 4–6). We consistently saw a slight increase in the level of ACT1 mRNA in the ΔNHP6 strain. (B) Nhp6 is not necessary for α2p repression. RNA was prepared from the depicted strains, and the Northern blot was probed with an MFA1 probe (Upper) or with an ACT1 probe (Lower). In lane 1, RNA was isolated from JD52, a MATa strain. In lane 2, RNA was isolated from JD53, which was used as wild-type parental MATα strain. Lane 3 contained RNA from JD53 lacking NHP6A and NHP6B (ΔNHP6). For lane 4, NHP6A and NHP6B had been reintegrated into the original loci (ΔNHP6 + NHP6). Lane 5 contained RNA from JD53 lacking TUP1 (ΔTUP1). (C) NHP6 and REG1 deletions are synthetically lethal. Shown are serial dilutions of the depicted S. cerevisiae strains carrying a URA3-marked Nhp6B expression plasmid (YCplac33-NHP6B) on medium lacking or containing FOA.
Figure 5
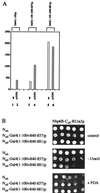
A truncated form of Gal4p, which displays an impaired interaction with Nhp6B, results in elevated levels of transcription upon deletion of NHP6. (A) Deleting NHP6 results in increased levels of transcription of a GAL1-LacZ reporter by a truncated form of Gal4p. Strains of the indicated genotype carrying a GAL1-LacZ reporter were transformed with the depicted expression plasmids. Arbitrary units of β-galactosidase activity are shown for the parental NLY2 strain, which lacks GAL4 and GAL80 in lanes 1, 3, and 5. The β-galactosidase activities of NLY2 cells additionally lacking NHP6A and NHP6B are shown in lanes 2, 4, and 6. Cells were grown in liquid glucose medium, and β-galactosidase activity was determined as described (33). Numbers were measured in triplicate, and standard deviations were less than 20%. All Gal4p derivatives were expressed from single-copy vectors. (B) Truncating the minimal activation domain of Gal4p results in decreased interaction with Nhp6B. Serial dilutions of cells coexpressing the depicted Nub and Cub fusions were grown on plates lacking tryptophan and leucine (Top), on plates additionally lacking uracil (Middle), or on plates containing FOA (Bottom). All proteins were expressed from single-copy vectors.
Similar articles
- Mutations in the yeast Nhp6 protein can differentially affect its in vivo functions.
Kruppa M, Kolodrubetz D. Kruppa M, et al. Biochem Biophys Res Commun. 2001 Feb 9;280(5):1292-9. doi: 10.1006/bbrc.2001.4273. Biochem Biophys Res Commun. 2001. PMID: 11162669 - Chromatin-mediated transcriptional regulation by the yeast architectural factors NHP6A and NHP6B.
Moreira JM, Holmberg S. Moreira JM, et al. EMBO J. 2000 Dec 15;19(24):6804-13. doi: 10.1093/emboj/19.24.6804. EMBO J. 2000. PMID: 11118215 Free PMC article. - SAGA is an essential in vivo target of the yeast acidic activator Gal4p.
Bhaumik SR, Green MR. Bhaumik SR, et al. Genes Dev. 2001 Aug 1;15(15):1935-45. doi: 10.1101/gad.911401. Genes Dev. 2001. PMID: 11485988 Free PMC article. - Bromodomain motifs and "scaffolding"?
Denis GV. Denis GV. Front Biosci. 2001 Sep 1;6:D1065-8. doi: 10.2741/A668. Front Biosci. 2001. PMID: 11532602 Free PMC article. Review. - Analysis of protein-protein proximities using the split-ubiquitin system.
Lehming N. Lehming N. Brief Funct Genomic Proteomic. 2002 Oct;1(3):230-8. doi: 10.1093/bfgp/1.3.230. Brief Funct Genomic Proteomic. 2002. PMID: 15239890 Review.
Cited by
- Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system.
Stynen B, Tournu H, Tavernier J, Van Dijck P. Stynen B, et al. Microbiol Mol Biol Rev. 2012 Jun;76(2):331-82. doi: 10.1128/MMBR.05021-11. Microbiol Mol Biol Rev. 2012. PMID: 22688816 Free PMC article. Review. - Biophysical characterization of DNA binding from single molecule force measurements.
Chaurasiya KR, Paramanathan T, McCauley MJ, Williams MC. Chaurasiya KR, et al. Phys Life Rev. 2010 Sep;7(3):299-341. doi: 10.1016/j.plrev.2010.06.001. Epub 2010 Jun 4. Phys Life Rev. 2010. PMID: 20576476 Free PMC article. Review. - Transcriptional activating regions target a cyclin-dependent kinase.
Ansari AZ, Koh SS, Zaman Z, Bongards C, Lehming N, Young RA, Ptashne M. Ansari AZ, et al. Proc Natl Acad Sci U S A. 2002 Nov 12;99(23):14706-9. doi: 10.1073/pnas.232573899. Epub 2002 Nov 4. Proc Natl Acad Sci U S A. 2002. PMID: 12417740 Free PMC article. - Dual binding modes for an HMG domain from human HMGB2 on DNA.
McCauley M, Hardwidge PR, Maher LJ 3rd, Williams MC. McCauley M, et al. Biophys J. 2005 Jul;89(1):353-64. doi: 10.1529/biophysj.104.052068. Epub 2005 Apr 15. Biophys J. 2005. PMID: 15833996 Free PMC article. - Eukaryotic HMGB proteins as replacements for HU in E. coli repression loop formation.
Becker NA, Kahn JD, Maher LJ 3rd. Becker NA, et al. Nucleic Acids Res. 2008 Jul;36(12):4009-21. doi: 10.1093/nar/gkn353. Epub 2008 May 31. Nucleic Acids Res. 2008. PMID: 18515834 Free PMC article.
References
- Wellhausen A, Lehming N. FEBS Lett. 1999;453:299–304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials