A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function - PubMed (original) (raw)
A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function
W Antonin et al. EMBO J. 2000.
Abstract
Sets of SNARE proteins mediate membrane fusion by assembling into core complexes. Multiple SNAREs are thought to function in different intracellular trafficking steps but it is often unclear which of the SNAREs cooperate in individual fusion reactions. We report that syntaxin 7, syntaxin 8, vti1b and endobrevin/VAMP-8 form a complex that functions in the fusion of late endosomes. Antibodies specific for each protein coprecipitate the complex, inhibit homotypic fusion of late endosomes in vitro and retard delivery of endocytosed epidermal growth factor to lysosomes. The purified proteins form core complexes with biochemical and biophysical properties remarkably similar to the neuronal core complex, although each of the four proteins carries a transmembrane domain and three have independently folded N-terminal domains. Substitution experiments, sequence and structural comparisons revealed that each protein occupies a unique position in the complex, with syntaxin 7 corresponding to syntaxin 1, and vti1b and syntaxin 8 corresponding to the N- and C-terminal domains of SNAP-25, respectively. We conclude that the structure of core complexes and their molecular mechanism in membrane fusion is highly conserved between distant SNAREs.
Figures
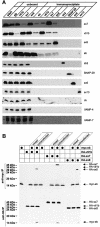
Fig. 1. Endobrevin, syntaxin 7, syntaxin 8 and vti1b form a complex in native membranes. (A) A complex of endobrevin, syntaxin 7, syntaxin 8 and vti1b co-immunoprecipitates from membrane extracts of rat liver. Triton X-100 solubilized membrane extracts were used for immunoprecipitations with antibodies specific for endobrevin (eb), vti1b, syntaxin 7 (sx7), syntaxin 8 (sx8), synaptobrevin 2 (sb2), SNAP-29, syntaxin 6 (sx6) and syntaxin 13 (sx13). The figure shows an immunoblot analysis of the starting material (start), unbound material obtained after precipitation (only for endobrevin, vti1b, syntaxin 7 and syntaxin 8) and immunoprecipitates. In each case, equal proportions of starting material, unbound material and immunoprecipitates were analyzed using antibodies specific for the indicated proteins. Cb, cellubrevin. (B) The complex of endobrevin, syntaxin 7, syntaxin 8 and vti1b exists in membranes before detergent solubilization. NRK cells were transfected in order to express myc-endobrevin, HA-vti1b, HA-syntaxin 7 and HA-syntaxin 8 either individually in different sets of cells, or to co-express HA-vti1b, HA-syntaxin 7 and HA-syntaxin 8 each together with myc-endobrevin (cotransfected). Membrane fractions prepared from each set of cells were extracted with Triton X-100 and used for immunoprecipitation with antibodies specific for the myc epitope (upper part) or the HA epitope (lower part). To check for complex formation after solubilization, aliquots of membranes obtained from individually transfected cells were mixed before detergent extraction (mixed). Note that coprecipitation of myc- and HA-tagged protein is only observed upon cotransfection.
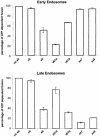
Fig. 2. Fab fragments specific for endobrevin, vti1b, syntaxin 7 and syntaxin 8 inhibit homotypic fusion of late endosomes. Homotypic fusion of early and late endosomes derived from PC12 cells was monitored in vitro using a content-mixing assay. Fab fragments (1.2 µM) specific for cellubrevin, endobrevin, vti1a, vti1b, syntaxin 7 and syntaxin 8 were used for pre-incubation of post-nuclear supernatants. ATP-dependent fusion activity in the absence of Fab fragments is defined as 100%. Values are given as means of three independent experiments, bars indicate the range. Fusion was initiated by mixing donor and acceptor fractions while simultaneously adding ATP and cytosol, followed by 30 min of incubation. See legend to Figure 1 for abbreviations.
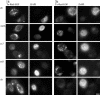
Fig. 3. Endocytic transport and lysosomal degradation of EGF is retarded by Fab fragments specific for endobrevin, vti1b, syntaxin 7 and syntaxin 8. HeLa cells were microinjected with Fab fragments specific for cellubrevin, endobrevin, vti1b, syntaxin 7 and syntaxin 8. EGF labeled with the fluorescent dye Texas Red was then added. After binding of EGF, endocytotic transport was allowed to proceed for 1 (left) or 3 h (right). At the end of the incubation, the cells were fixed and analyzed by fluorescence microscopy. Microinjected cells were identified by the nuclear staining by co-injected DAPI. Bar, 10 µm.
Fig. 4. Biochemical characterization of the endosomal SNARE complex. (A) Recombinant endobrevin, syntaxin 7, syntaxin 8 and vti1b (each lacking their transmembrane domain) form a partially SDS-resistant complex. After assembly, the complex was purified using ion exchange chromatography and analyzed by SDS–PAGE, with or without boiling in SDS sample buffer. The asterisk denotes a contaminating breakdown product of syntaxin 8. (B) Limited proteolysis of the cytoplasmic complex gives rise to a partially SDS-resistant core complex. The complex shown in (A) was treated with trypsin for 30 min. The reaction mixture was separated by size-exclusion chromatography, and the protein-containing fractions were analyzed by SDS–PAGE with or without boiling in SDS sample buffer. The arrowhead denotes the position of the protease-resistant core complex in the non-boiled sample. All N-termini were identified by sequencing. Note that a similar pattern was obtained when proteinase K or chymotrypsin (not shown) was used. (C) Cytoplasmic and core complexes are defined particles with a 1:1:1:1 stoichiometry. Cytoplasmic complexes (upper panel) and core complexes (lower panel) were analyzed by size-exclusion chromatography. Absorption was measured at 280 nm (continuous lines, left axis), and molecular masses were determined by multi-angle laser light scattering (broken lines, right axis). Separations were carried out in either low salt buffer (20 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT; black lines) or high salt buffer (1 M NaCl instead of 150 mM; grey lines). Protein concentrations of the injected samples were 10 µM.
Fig. 5. Disassembly and assembly of endosomal SNARE complexes. (A) Disassembly by NSF and α-SNAP of the cytoplasmic complex and the core complex. Purified cytoplasmic complexes and core complexes were incubated as indicated in the presence of ATP. NSF-driven disassembly was monitored by the disappearance of the SDS-resistant complexes using an endobrevin-specific antiserum specific for their detection after immunoblotting. (B) Fab fragments specific for the endosomal SNARE proteins inhibit assembly of the core complex. SNARE motifs of endobrevin, vti1b, syntaxin 7 and syntaxin 8 were each incubated with two different concentrations of the respective Fab fragments. The other three SNARE motifs were then added to allow formation of core complexes that were determined as in (A). For control, endobrevin was incubated with Fab fragments specific for cellubrevin. SDS-resistant core complexes are dissociated by boiling (left).
Fig. 6. Structural characterization of endosomal SNARE motifs and their interactions. (A) CD spectra of purified SNARE motifs of endobrevin, vti1b, syntaxin 7 and syntaxin 8, and of an equimolar mixture of all four. CD was recorded in 40 mM sodium phosphate at 25°C at a protein concentration of 15 µM. All samples were incubated overnight at 4°C prior to spectroscopy. (B) Changes in the mean residue ellipticity (Θ) at 220 nm induced by interaction between SNARE motifs of endobrevin, vti1b, syntaxin 7 and syntaxin 8. CD was recorded in buffer containing 40 mM sodium phosphate at 25°C at a protein concentration of 15 µM. White columns represent theoretically non-interacting mean residue ellipticity at 220 nm calculated from the observed CD spectra of the individual proteins (columns 1–4). To obtain the observed CD spectra (black columns) of the various combinations, the proteins were incubated overnight at 4°C using equimolar ratios. (C) Percentage of α-helical structure of the purified core complex as a function of temperature. Change in the ellipticity at 220 nm was monitored and the α-helical content was calculated.
Fig. 7. SDS-resistant complexes between neuronal and endosomal SNAREs form only when equivalent SNARE motifs are exchanged. Combinations of purified SNARE motifs were mixed as indicated in approximately equimolar ratios, incubated overnight, and analyzed by SDS–PAGE without boiling and staining with Coomassie Blue.
Fig. 8. Sequence alignment of the SNARE motifs of different Q-SNAREs. Shading denotes the position of residues that in the crystal structure of the neuronal SNARE complex form interacting layers of the four associating α-helices [numbered according to Fasshauer et al. (1998b)]. Side chains that differentiate between syntaxin, SNAP-25/N-terminal domain (SN1) and SNAP-25/C-terminal domain (SN2) subfamily members are boxed. DDBJ/EMBL/GenBank accession Nos are: sx1, P3285; sx2, L20823; sx3, Q08849; sx4, Q08850; sx5, Q08851; sx7, AF031430; sx13, AF031430; sx6, U56815; sx8, AF033109; SNAP-23, AF052596; SNAP-25, AF245227; SNAP-29, AF260577; vti1a, AF035209; and vti1b, AF035208.
Fig. 9. Cartoon showing a comparison of the neuronal SNARE complex and the endosomal SNARE complex. TMR, transmembrane region. See text for details. Note that the connection between the N-terminal domains and the helical SNARE motifs may be structured and that the N-terminal domains may fold back and interact with the helical bundle in the endosomal complex. Also, note that it is not known whether the N-terminus of syntaxin 7 forms a trihelical bundle like that of syntaxin 1.
Similar articles
- Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs.
Antonin W, Fasshauer D, Becker S, Jahn R, Schneider TR. Antonin W, et al. Nat Struct Biol. 2002 Feb;9(2):107-11. doi: 10.1038/nsb746. Nat Struct Biol. 2002. PMID: 11786915 - Syntaxin 7, syntaxin 8, Vti1 and VAMP7 (vesicle-associated membrane protein 7) form an active SNARE complex for early macropinocytic compartment fusion in Dictyostelium discoideum.
Bogdanovic A, Bennett N, Kieffer S, Louwagie M, Morio T, Garin J, Satre M, Bruckert F. Bogdanovic A, et al. Biochem J. 2002 Nov 15;368(Pt 1):29-39. doi: 10.1042/BJ20020845. Biochem J. 2002. PMID: 12175335 Free PMC article. - Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly.
Fasshauer D, Eliason WK, Brünger AT, Jahn R. Fasshauer D, et al. Biochemistry. 1998 Jul 21;37(29):10354-62. doi: 10.1021/bi980542h. Biochemistry. 1998. PMID: 9671503 - Phylogenetic analysis of membrane trafficking proteins: a family reunion and secondary structure predictions.
Terrian DM, White MK. Terrian DM, et al. Eur J Cell Biol. 1997 Jul;73(3):198-204. Eur J Cell Biol. 1997. PMID: 9243180 Review. - Protein interactions implicated in neurotransmitter release.
el Far O, O'Connor V, Dresbach T, Pellegrini L, DeBello W, Schweizer F, Augustine G, Heuss C, Schäfer T, Charlton MP, Betz H. el Far O, et al. J Physiol Paris. 1998 Apr;92(2):129-33. doi: 10.1016/S0928-4257(98)80150-2. J Physiol Paris. 1998. PMID: 9782456 Review.
Cited by
- Phosphoproteomic Analysis of Rat Neutrophils Shows the Effect of Intestinal Ischemia/Reperfusion and Preconditioning on Kinases and Phosphatases.
Tahir M, Arshid S, Fontes B, S Castro M, Sidoli S, Schwämmle V, Luz IS, Roepstorff P, Fontes W. Tahir M, et al. Int J Mol Sci. 2020 Aug 13;21(16):5799. doi: 10.3390/ijms21165799. Int J Mol Sci. 2020. PMID: 32823483 Free PMC article. - Three-dimensional structure of the amino-terminal domain of syntaxin 6, a SNAP-25 C homolog.
Misura KM, Bock JB, Gonzalez LC Jr, Scheller RH, Weis WI. Misura KM, et al. Proc Natl Acad Sci U S A. 2002 Jul 9;99(14):9184-9. doi: 10.1073/pnas.132274599. Epub 2002 Jun 24. Proc Natl Acad Sci U S A. 2002. PMID: 12082176 Free PMC article. - OsSYP121 Accumulates at Fungal Penetration Sites and Mediates Host Resistance to Rice Blast.
Cao WL, Yu Y, Li MY, Luo J, Wang RS, Tang HJ, Huang J, Wang JF, Zhang HS, Bao YM. Cao WL, et al. Plant Physiol. 2019 Apr;179(4):1330-1342. doi: 10.1104/pp.18.01013. Epub 2019 Jan 7. Plant Physiol. 2019. PMID: 30617050 Free PMC article. - Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell.
Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. Sanderfoot AA, et al. Mol Biol Cell. 2001 Dec;12(12):3733-43. doi: 10.1091/mbc.12.12.3733. Mol Biol Cell. 2001. PMID: 11739776 Free PMC article. - D53 is a novel endosomal SNARE-binding protein that enhances interaction of syntaxin 1 with the synaptobrevin 2 complex in vitro.
Proux-Gillardeaux V, Galli T, Callebaut I, Mikhailik A, Calothy G, Marx M. Proux-Gillardeaux V, et al. Biochem J. 2003 Feb 15;370(Pt 1):213-21. doi: 10.1042/BJ20021309. Biochem J. 2003. PMID: 12376003 Free PMC article.
References
- Advani R.J., Bae,H.R., Bock,J.B., Chao,D.S., Doung,Y.C., Prekeris,R., Yoo,J.S. and Scheller,R.H. (1998) Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J. Biol. Chem., 273, 10317–10324. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases