She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA - PubMed (original) (raw)
She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA
R M Long et al. EMBO J. 2000.
Abstract
In Saccharomyces cerevisiae, Ash1p is a specific repressor of transcription that localizes exclusively to daughter cell nuclei through the asymmetric localization of ASH1 mRNA. This localization requires four cis-acting localization elements located in the ASH1 mRNA, five trans-acting factors, one of which is a myosin, and the actin cytoskeleton. The RNA-binding proteins that interact with these cis-elements remained to be identified. Starting with the 3' most localization element of ASH1 mRNA in the three-hybrid assay, element E3, we isolated a clone corresponding to the C-terminus of She3p. We also found that She3p and She2p interact, and this interaction is essential for the binding of She3p with element E3 in vivo. Moreover, She2p was observed to bind the E3 RNA directly in vitro and each of the ASH1 cis-acting localization elements requires She2p for their localization function. By tethering a She3p-MS2 fusion protein to a reporter RNA containing MS2 binding sites, we observed that She2p is dispensable for She3p-MS2-dependent RNA localization.
Figures
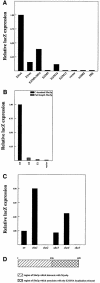
Fig. 1. By three-hybrid analysis, the C-terminal section of She3p preferentially interacts with the E3 ASH1 RNA localization element. (A) Strain YBZ-1 transformed with a plasmid expressing Gal4p-AD fused to the C-terminus of She3p along with a plasmid expressing one of the indicated MS2 fusion RNAs. Liquid cultures were grown for each strain, and the cells harvested by centrifugation. The cells were permeabilized and β-galactosidase expression levels were determined. β-galactosidase expression levels were normalized to cells expressing the MS2–E3 fusion RNA. (B) Strain YBZ-1 transformed with either a plasmid expressing the C-terminus of She3p fused to Gal4p-AD or full-length She3p fused to Gal4p-AD along with plasmids expressing the indicated MS2 fusion RNAs. Liquid cultures were grown for each strain and used for β-galactosidase assays. β-galactosidase expression levels were normalized to cells expressing the She3p C-terminus fused to Gal4p-AD and the MS2–E3 fusion RNA. (C) Yeast strain YBZ-1 deleted of myo4, she2, she3, she4 or she5. The strains were subsequently transformed with a plasmid expressing full-length She3p fused to Gal4p-AD along with a plasmid expressing the MS2–E3 fusion RNA. Transformants were grown in liquid culture and used for β-galactosidase assays. β-galactosidase levels were normalized to the wild-type strain. (D) Schematic representation of She3p. Amino acids 1–236 of She3p contain a domain required for the association with Myo4p, while amino acids 236–425 contain a domain required for the in vivo association with the E3 ASH1 RNA localization element.
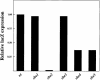
Fig. 2. She3p C-terminal-dependent lacZ expression assayed by three-hybrid analysis is dependent on She2p. Yeast strain YBZ-1 deleted of myo4, she2, she3, she4 and she5. These strains were subsequently transformed with a plasmid expressing the C-terminus She3p fused to Gal4p-AD and MS2–E3 fusion RNA. The transformants were grown in liquid culture and processed for β-galactosidase assays. β-galactosidase expression levels were normalized to the wild-type strain.
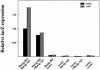
Fig. 3. By the two-hybrid assay, She2p and She3p interact. Yeast strains PJ69-4a (ASH1) and YLM445 (ash1) were transformed in combinations with the following plasmids: She2p fused to Gal4p-BD (She2p–BD), full-length She3p fused to Gal4p-AD (She3p–AD), the C-terminus of She3p fused to Gal4p-AD (cShe3p–AD), the vector corresponding to Gal4p-BD (pGBDU-c2) or the vector corresponding to Gal4p-AD (pACT2). Transformants were grown in liquid culture and processed for β-galactosidase assays. β-galactosidase expression levels were normalized to strain PJ69-4a transformed with the plasmid expressing She2p fused to Gal4p-BD and the plasmid expressing full-length She3p fused to Gal4p-AD.
Fig. 4. She2p RNA-binding activity as assayed by a UV cross-linking assay. (A) One nanogram of 32P-labeled E3 RNA (lanes 2 and 4) or pGEM-4Z RNA (lanes 1 and 3) was incubated with 1.0 µg of purified GST–She2p (lanes 3 and 4) or GST (lanes1 and 2) corresponding to an equivalent volume of GST–She2p used for UV cross-linking. The GST–She2p–E3 complex migrates at 49.6 kDa. The arrow indicates the position of the GST–She2p–E3 complex. (B) Specific cross-linking of GST–She2p to E3. UV cross-linking was performed with GST–She2p and 32P-labeled E3 in the absence (lanes 1 and 5) or presence of 100× (lane 2), 500× (lane 3) and 1000× (lane 4) pGEM-4Z RNA, as well as 100× (lane 6), 500× (lane 7) and 1000× (lane 8) E3 RNA. (C) Competition of GST–She2p–E3 UV cross-linked product with mutant E3 elements. GST–She2p was incubated with 32P-labeled E3 in the absence of pGEM and E3 RNA (lane 1) or 1000× pGEM RNA (lane 2), 1000× pGEM/1000× E3 (lane3), 1000× pGEM/1000× E3/M9 (lane 4), 1000× pGEM/1000× E3/M9 + M9A (lane 5), 1000× pGEM/1000× E3/M13 (lane 6) or 1000× pGEM/1000× E3/M14 (lane 7). (D) Competition of GST–She2p–E3 UV cross-linked product with _ASH1 cis_-acting localization elements. GST–She2p was incubated with 32P-labeled E3 in the absence of pGEM and E3 RNA (lane 1), 1000× pGEM RNA (lane 2), 1000× pGEM/1000× E1 (lane3), 1000× pGEM/1000× E2A (lane 4), 1000× pGEM/1000× E2B (lane 5) or 1000× pGEM/1000× E3 (lane 6).
Fig. 5. RNA localization activity for each of the _cis_-acting localization elements is dependent on She2p. (A) Wild-type (wt) and she2 yeast strains were transformed with a plasmid containing the galactose-inducible lacZ-E3 cassette, and the cells were processed for fluorescent in situ hybridization (FISH). (B) Fraction of budding cells with localized lacZ mRNA for each of the _cis_-acting localization elements in wild-type and she2 cells. Cells expressing each of the reporter constructs (lacZ-E1, lacZ-E2A, lacZ-E2B and lacZ-E3) were grown in galactose-containing media and processed for in situ hybridization. LacZ RNA was categorized as localized or delocalized from cells displaying FISH signal.
Fig. 5. RNA localization activity for each of the _cis_-acting localization elements is dependent on She2p. (A) Wild-type (wt) and she2 yeast strains were transformed with a plasmid containing the galactose-inducible lacZ-E3 cassette, and the cells were processed for fluorescent in situ hybridization (FISH). (B) Fraction of budding cells with localized lacZ mRNA for each of the _cis_-acting localization elements in wild-type and she2 cells. Cells expressing each of the reporter constructs (lacZ-E1, lacZ-E2A, lacZ-E2B and lacZ-E3) were grown in galactose-containing media and processed for in situ hybridization. LacZ RNA was categorized as localized or delocalized from cells displaying FISH signal.
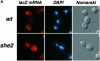
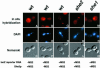
Fig. 6. RNA localization directed by She3p–MS2 is independent of She2p. (A–C) Representative images for in situ hybridization to lacZ mRNA, DAPI staining and Nomarski optics of wild-type cells expressing the She3p–MS2 fusion protein and the lacZ reporter RNA devoid of MS2 binding sites. (D–F) Images for wild-type cells expressing She3p and the lacZ-MS2 reporter RNA. (G–I) Images for wild-type cells expressing the She3p–MS2 fusion protein and the lacZ-MS2 reporter mRNA. Images for she2 (J–L) and she1 (M–O) cells, respectively, expressing the She3p–MS2 fusion protein and the lacZ-MS2 reporter mRNA.
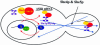
Fig. 7. Model for the localization of ASH1 mRNA. In the nucleus, ASH1 mRNA is identified by Loc1p in association with nucleocytoplasmic shuttling proteins. Following transport through the nuclear pores, ASH1 mRNA is rapidly assembled into a transport particle through the RNA-binding activity of She2p. The She2p–ASH1 mRNA complex associates with Myo4p–She3p to transport ASH1 mRNA to the bud tip by movement on actin cables. Once at the bud tip, ASH1 mRNA will be anchored through a mechanism that remains to be determined.
Similar articles
- She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p.
Böhl F, Kruse C, Frank A, Ferring D, Jansen RP. Böhl F, et al. EMBO J. 2000 Oct 16;19(20):5514-24. doi: 10.1093/emboj/19.20.5514. EMBO J. 2000. PMID: 11032818 Free PMC article. - ASH1 mRNA anchoring requires reorganization of the Myo4p-She3p-She2p transport complex.
Gonsalvez GB, Little JL, Long RM. Gonsalvez GB, et al. J Biol Chem. 2004 Oct 29;279(44):46286-94. doi: 10.1074/jbc.M406086200. Epub 2004 Aug 23. J Biol Chem. 2004. PMID: 15328357 - The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p.
Takizawa PA, Vale RD. Takizawa PA, et al. Proc Natl Acad Sci U S A. 2000 May 9;97(10):5273-8. doi: 10.1073/pnas.080585897. Proc Natl Acad Sci U S A. 2000. PMID: 10792032 Free PMC article. - RNA localization in yeast: moving towards a mechanism.
Gonsalvez GB, Urbinati CR, Long RM. Gonsalvez GB, et al. Biol Cell. 2005 Jan;97(1):75-86. doi: 10.1042/BC20040066. Biol Cell. 2005. PMID: 15601259 Review. - RNA localization: SHEdding light on the RNA-motor linkage.
Kwon S, Schnapp BJ. Kwon S, et al. Curr Biol. 2001 Mar 6;11(5):R166-8. doi: 10.1016/s0960-9822(01)00084-7. Curr Biol. 2001. PMID: 11267883 Review.
Cited by
- Inducible control of subcellular RNA localization using a synthetic protein-RNA aptamer interaction.
Belmont BJ, Niles JC. Belmont BJ, et al. PLoS One. 2012;7(10):e46868. doi: 10.1371/journal.pone.0046868. Epub 2012 Oct 8. PLoS One. 2012. PMID: 23056498 Free PMC article. - A novel mRNA affinity purification technique for the identification of interacting proteins and transcripts in ribonucleoprotein complexes.
Slobodin B, Gerst JE. Slobodin B, et al. RNA. 2010 Nov;16(11):2277-90. doi: 10.1261/rna.2091710. Epub 2010 Sep 28. RNA. 2010. PMID: 20876833 Free PMC article. - Mechanisms and consequences of subcellular RNA localization across diverse cell types.
Engel KL, Arora A, Goering R, Lo HG, Taliaferro JM. Engel KL, et al. Traffic. 2020 Jun;21(6):404-418. doi: 10.1111/tra.12730. Epub 2020 Apr 29. Traffic. 2020. PMID: 32291836 Free PMC article. Review. - Daughter cell-targeted mRNAs can achieve segregation without the universal Endoplasmic Reticulum docker She2p.
Samardak K, Moriel-Carretero M. Samardak K, et al. MicroPubl Biol. 2021 Sep 13;2021:10.17912/micropub.biology.000458. doi: 10.17912/micropub.biology.000458. eCollection 2021. MicroPubl Biol. 2021. PMID: 34532701 Free PMC article. - Walking the line: mechanisms underlying directional mRNA transport and localisation in neurons and beyond.
Abouward R, Schiavo G. Abouward R, et al. Cell Mol Life Sci. 2021 Mar;78(6):2665-2681. doi: 10.1007/s00018-020-03724-3. Epub 2020 Dec 20. Cell Mol Life Sci. 2021. PMID: 33341920 Free PMC article. Review.
References
- Beach D.L., Salmon,E.D. and Bloom,K. (1999) Localization and anchoring of mRNA in budding yeast. Curr. Biol., 9, 569–578. - PubMed
- Bertrand E., Chartrand,P., Schaefer,M., Shenoy,S.M., Singer,R.H. and Long,R.M. (1998) Localization of ASH1 mRNA particles in living yeast. Mol. Cell, 2, 437–445. - PubMed
- Bobola N., Jansen,R.-P., Shin,T.H. and Nasmyth,K. (1996) Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell, 84, 699–709. - PubMed
- Chartrand P., Meng X., Singer,R.H. and Long,R.M. (1999) Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol., 9, 333–336. - PubMed
- Evangelista M., Blundell,K., Longtine,M.S., Chow,C.J., Adames,N., Pringle,J.R., Peter,M. and Boone,C. (1997) Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science, 276, 118–122. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2-T32-CA09475/CA/NCI NIH HHS/United States
- T32 CA009475/CA/NCI NIH HHS/United States
- R01 GM057071/GM/NIGMS NIH HHS/United States
- GM57071/GM/NIGMS NIH HHS/United States
- GM60392/GM/NIGMS NIH HHS/United States
- R01 GM060392/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases