Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores - PubMed (original) (raw)
Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores
H Scherthan et al. Mol Biol Cell. 2000 Dec.
Free PMC article
Abstract
Mammalian telomeres consist of TTAGGG repeats, telomeric repeat binding factor (TRF), and other proteins, resulting in a protective structure at chromosome ends. Although structure and function of the somatic telomeric complex has been elucidated in some detail, the protein composition of mammalian meiotic telomeres is undetermined. Here we show, by indirect immunofluorescence (IF), that the meiotic telomere complex is similar to its somatic counterpart and contains significant amounts of TRF1, TRF2, and hRap1, while tankyrase, a poly-(ADP-ribose)polymerase at somatic telomeres and nuclear pores, forms small signals at ends of human meiotic chromosome cores. Analysis of rodent spermatocytes reveals Trf1 at mouse, TRF2 at rat, and mammalian Rap1 at meiotic telomeres of both rodents. Moreover, we demonstrate that telomere repositioning during meiotic prophase occurs in sectors of the nuclear envelope that are distinct from nuclear pore-dense areas. The latter form during preleptotene/leptotene and are present during entire prophase I.
Figures
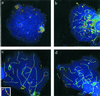
Figure 1
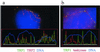
Spreads of human spermatocytes I immunostained with anti-TRF1 (#371; Cy3, red) and anti-SCP3 synaptonemal complex protein (FITC, green). (a) Spread preleptotene nucleus (identified by its extensive diameter and week patchy SCP3 deposits to the right), which exhibits distinct TRF1 signal spots that equal the diploid number of individual telomeres (92). (b) Mildly spread leptotene nucleus (focal plane on top of nucleus) with TRF1 signals at the ends of developing axial elements (green threads), which are accumulated at a limited region of the nucleus (bouquet arrangement). (c) Pachytene spread. Distinct TRF1 signals are seen at both ends of the SCs. Note that TRF1 forms thread-like connections between several nonhomologous telomeres. The enlarged inset displays two nonhomologous SC ends that are connected by a TRF1 stretch. (d) Late pachytene spread: prominent TRF1 signals cap the SC ends. The sex chromosome cores form a condensed XY bivalent (arrow) that also contains TRF1 at its telomeres. DNA is counterstained with DAPI (blue).
Figure 2
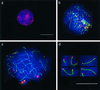
(mTrf1) Spread pachytene spermatocyte I of the mouse (Mus musculus) stained with anti-SCP3 antiserum (FITC, green) and anti-mouse Trf1 (Cy3; red). Strong Trf1 signals are present at ends of SCs, unpaired axes (arrow), and the telomeres of the sex chromosomes (arrowhead). One strong Trf1 signal is seen at the association site of the distal ends of the XY pair. (mRap1) IF staining of mammalian Rap1 with anti-hRap1 Abs (red, Cy3) in a spread mouse pachytene nucleus. The ends of the SCs are capped by strong, large mRap1 signals, which sometimes extend beyond the SC ends. The XY pair displays mRap1 signals at the ends of the paired and the two unpaired meiotic chromosome cores (arrowhead). The fuzzy green material at three bivalents results from SCP3 fluorescence at remnants of nucleolar material carried by three autosomes. DNA is shown in blue (DAPI).
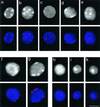
Figure 3
Telomere redistribution during mouse spermatogenesis as displayed by Trf1 immunofluorescence (Cy3, red). Staging was deduced from stage-specific heterochromatin and telomere distribution patterns (see Scherthan et al., 1996). DAPI staining of nuclear DNA is given at the top (gray). DAPI-bright heterochromatin is seen as whitish clusters. (a) Nucleus of a spermatogonium with dispersed telomere distribution. (b) Premeiotic nucleus with prominent intranuclear heterochromatin clusters and dispersed telomeric Trf1 signals. (c) Preleptotene nucleus with faint peripheral heterochromatin and scattered Trf1 telomere signals (d) Late preleptotene nucleus displaying exclusively perinuclear telomere distribution. Note the small telomere signals, compared with f and g. (e) Top of bouquet nucleus (corresponding to leptotene/zygotene) with clustered telomeres. Small Trf1 signal diameter indicates that most telomeres have not yet paired. (f and g) Pachytene nuclei as identified by prominent peripheral heterochromatin clusters and large peripheral telomere signals. (f) Top of pachytene nucleus with numerous signals. (g) Focal plane at maximum diameter of a pachytene nucleus displaying Trf1 signals at the nuclear envelope. (h-k) Haploid round spermatids showing Trf1 telomere signals at the DAPI-bright heterochromatin chromocenters over the central region of the spermatid nuclei. Bar: 10 μm, it applies to all details.
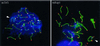
Figure 4
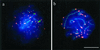
Spread human spermatocytes I after immunostaining with anti-TRF2 (Cy3; red) and anti-SCP3 (FITC, green). (a) Top region of a mildly spread leptotene nucleus, which shows distinct TRF2 signal spots at the ends of developing axial elements (faint green threads). Ends of cores and TRF2 signals overlap (yellow) (b) Focal plane at the top of a spread pachytene nucleus. Distinct TRF2 signals are seen at the ends of the SCs. Most of the SCs extend beyond the focal plane. DNA is stained with DAPI (blue). Bar represents 10 μm.
Figure 5
Analysis of colocalization of fluorescent signals of telomeric proteins in human testes nuclei. Fluorescence profile analysis is conducted along polygon lines drawn across telomeric signals and adjacent chromatin in raw digital images (see methods). Colocalization is indicated by signal peaks in the green and red channel at identical positions. (a) A fluorescence profile derived from a line polygon drawn across 19 telomeres at the top of a bouquet nucleus (leptotene/zygotene) costained with TRF1 (green, FITC) and TRF2 (red, Cy3). Perfect colocalization of similar sized signals is seen. The DAPI profile (DNA, blue) appears uneven since it runs along the nuclear periphery. (b) Colocalization of tankyrase (red) and five TRF1 signals in a sector of a spread spermatocyte nucleus. Each TRF1 signal peak is colocalized with a distinct tankyrase signal peak. Additional tankyrase signals which create additional peaks in the red channel likely result from reminders of nontelomeric tankyrase_e.g._, at nuclear pore remnants (see text).
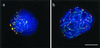
Figure 6
Ionic detergent spread human testicular nuclei after immunostaining with anti-tankyrase (Cy3; red) and anti-SCP3 (FITC, green) antibodies. DNA is stained with DAPI (blue). (a) Premeiotic cell displaying numerous tankyrase signals scattered throughout the nucleus and its periphery. This pattern agrees with the localization of tankyrase at telomeres and nuclear pores (Smith and de Lange 1999). (b) Top of a mildly spread leptotene nucleus at the bouquet stage (accumulated telomeres). The axial elements (green threads) are capped with tankyrase signals (reddish). Surplus small red signal dots are scattered between the clustered chromosome ends and possibly represent remnants of extratelomeric tankyrase. (c) Pachytene spread showing small but distinct tankyrase signals at the ends of the SCs (green). The bright red dots are background grains often seen in this testes sample. (d) Magnified details of SCs showing distinct tankyrase signals at their termini. Sometimes, the tankyrase signals were split in two and seen to extend beyond the SC ends. This is particularly evident in the first two details. In the upper left detail the axial core extends beyond the focal plane. Bars in a and d represent 10 μm. The bar in a applies also to b and c.
Figure 7
Immunofluorescent staining of hRap1 (red, Cy3) in mildly spread human spermatocytes I. Focal planes at top of nuclei. (a) A human leptotene spermatocyte displays small but distinct hRap1 signals at the ends of unpaired chromosome axes (green, FITC). (b) Large hRap1 signals are seen at the ends of SCs of a mildly spread pachytene nucleus. DNA is counterstained in blue (DAPI). Bar: 10 μm.
Figure 8
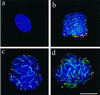
Mildly spread nuclei from testicular suspensions of the rat (Rattus norvegicus) immunostained with anti-hRap1 Abs (Cy3; red) and anti-SCP3 Abs (FITC, green). DNA is shown in blue (DAPI). (a) Premeiotic nucleus with numerous mRap1 signal spots. SC proteins are absent. (b) Mildly spread leptotene/early zygotene nucleus. Numerous thin axial cores are seen with synapsis in progress between two chromosome ends (thick green signal stretch) near the bouquet basis. The latter contains numerous closely spaced chromosome ends, capped with strong mRap1 signals. (c) Mildly spread late pachytene nucleus with distinct mRap1 signals at SC ends. Some SCs extend beyond the focal plane, which is at the top of this nucleus. (d) Mildly spread pachytene nucleus with strong mRap1 signals at SC ends. The closely spaced ends of the backfolded cores of the XY bivalent exhibit distinct mRap1 signals (arrowhead). Top of nucleus shown. Bar in c: 10 μm.
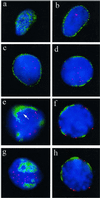
Figure 9
Distribution of TRF1-tagged telomeres (red, Cy3) in relation to nuclear pore (NUP) positioning (green, FITC; detected with mAb414) in undisrupted human premeiotic and meiotic testicular suspension nuclei. (a) Focal plane at the top of a premeiotic nucleus reveals the typical, more or less even distribution of nuclear pores. (b) Nuclear pore (NUP) IF at the equatorial plane of the same nucleus creates a rim-like signal distribution, while a few telomere signals are present in the interior. (c) NUP rim-staining at the equator of another premeiotic nucleus. Telomere signals are seen in the nuclear interior. (d) Patchy perinuclear distribution of NUPs of an early meiotic nucleus (as deduced by its increased DAPI intensity) with internal telomeres (preleptotene). (e) Focal plane at the top of a spermatocyte I nucleus that displays pronounced accumulation of NUPs. Telomeres are at the nuclear periphery. The immediate vicinity of a few telomeres that are close to NUP clusters is devoid of NUP fluorescence (arrow). (f) Center view of a pachytene nucleus (note the thread-like DAPI-stained chromatin). Telomeres locate adjacent to NUP patches or at NUP-free regions of the nuclear envelope. (g and h) Top and equatorial focal plane, respectively, of an early pachytene nucleus (deduced from locally accumulated telomeres and thread-like chromatin) with telomeres remote from NPC-dense areas. In the equatorial plane, a telomere spot colocalizes with NUP signal stretch (h, to the right). Nuclear DNA is counterstained in blue (DAPI).
Similar articles
- Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas.
Matsutani N, Yokozaki H, Tahara E, Tahara H, Kuniyasu H, Haruma K, Chayama K, Yasui W, Tahara E. Matsutani N, et al. Int J Oncol. 2001 Sep;19(3):507-12. Int J Oncol. 2001. PMID: 11494028 - Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres.
Cook BD, Dynek JN, Chang W, Shostak G, Smith S. Cook BD, et al. Mol Cell Biol. 2002 Jan;22(1):332-42. doi: 10.1128/MCB.22.1.332-342.2002. Mol Cell Biol. 2002. PMID: 11739745 Free PMC article. - Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes.
Smith S, de Lange T. Smith S, et al. J Cell Sci. 1999 Nov;112 ( Pt 21):3649-56. doi: 10.1242/jcs.112.21.3649. J Cell Sci. 1999. PMID: 10523501 - Telomere Repeat-Binding Factor 2 Is Responsible for the Telomere Attachment to the Nuclear Membrane.
Ilicheva NV, Podgornaya OI, Voronin AP. Ilicheva NV, et al. Adv Protein Chem Struct Biol. 2015;101:67-96. doi: 10.1016/bs.apcsb.2015.06.009. Epub 2015 Oct 21. Adv Protein Chem Struct Biol. 2015. PMID: 26572976 Review. - Telomere repeat binding factors: keeping the ends in check.
Karlseder J. Karlseder J. Cancer Lett. 2003 May 15;194(2):189-97. doi: 10.1016/s0304-3835(02)00706-1. Cancer Lett. 2003. PMID: 12757977 Review.
Cited by
- The SUN1-SPDYA interaction plays an essential role in meiosis prophase I.
Chen Y, Wang Y, Chen J, Zuo W, Fan Y, Huang S, Liu Y, Chen G, Li Q, Li J, Wu J, Bian Q, Huang C, Lei M. Chen Y, et al. Nat Commun. 2021 May 26;12(1):3176. doi: 10.1038/s41467-021-23550-w. Nat Commun. 2021. PMID: 34039995 Free PMC article. - Telomere attachment, meiotic chromosome condensation, pairing, and bouquet stage duration are modified in spermatocytes lacking axial elements.
Liebe B, Alsheimer M, Höög C, Benavente R, Scherthan H. Liebe B, et al. Mol Biol Cell. 2004 Feb;15(2):827-37. doi: 10.1091/mbc.e03-07-0524. Epub 2003 Dec 2. Mol Biol Cell. 2004. PMID: 14657244 Free PMC article. - A Novel Meiosis-Related lncRNA, Rbakdn, Contributes to Spermatogenesis by Stabilizing Ptbp2.
Liu W, Zhao Y, Liu X, Zhang X, Ding J, Li Y, Tian Y, Wang H, Liu W, Lu Z. Liu W, et al. Front Genet. 2021 Oct 11;12:752495. doi: 10.3389/fgene.2021.752495. eCollection 2021. Front Genet. 2021. PMID: 34707642 Free PMC article. - Chromosome Dynamics Regulating Genomic Dispersion and Alteration of Nucleolus Organizer Regions (NORs).
Hirai H. Hirai H. Cells. 2020 Apr 15;9(4):971. doi: 10.3390/cells9040971. Cells. 2020. PMID: 32326514 Free PMC article. Review. - Role for telomere cap structure in meiosis.
Maddar H, Ratzkovsky N, Krauskopf A. Maddar H, et al. Mol Biol Cell. 2001 Oct;12(10):3191-203. doi: 10.1091/mbc.12.10.3191. Mol Biol Cell. 2001. PMID: 11598202 Free PMC article.
References
- Albini SM, Jones GH. Synaptonemal complex-associated centromeres and recombination nodules in plant meiocytes prepared by an improved surface-spreading technique. Exp Cell Res. 1984;155:588–592. - PubMed
- Bennett V. Ankyrins. Adaptors between diverse plasma membrane proteins and the cytoplasm. J Biol Chem. 1992;267:8703–8706. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous