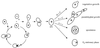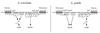Signal transduction cascades regulating fungal development and virulence - PubMed (original) (raw)
Review
Signal transduction cascades regulating fungal development and virulence
K B Lengeler et al. Microbiol Mol Biol Rev. 2000 Dec.
Abstract
Cellular differentiation, mating, and filamentous growth are regulated in many fungi by environmental and nutritional signals. For example, in response to nitrogen limitation, diploid cells of the yeast Saccharomyces cerevisiae undergo a dimorphic transition to filamentous growth referred to as pseudohyphal differentiation. Yeast filamentous growth is regulated, in part, by two conserved signal transduction cascades: a mitogen-activated protein kinase cascade and a G-protein regulated cyclic AMP signaling pathway. Related signaling cascades play an analogous role in regulating mating and virulence in the plant fungal pathogen Ustilago maydis and the human fungal pathogens Cryptococcus neoformans and Candida albicans. We review here studies on the signaling cascades that regulate development of these and other fungi. This analysis illustrates both how the model yeast S. cerevisiae can serve as a paradigm for signaling in other organisms and also how studies in other fungi provide insights into conserved signaling pathways that operate in many divergent organisms.
Figures
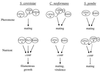
FIG. 1
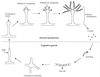
Life cycle of S. cerevisiae. Haploid yeast cells mate on rich medium. Diploid yeast cells adopt alternative fates depending on the availability of nutrients. N + C, abundant nitrogen and fermentable carbon source; n + C, limiting nitrogen and abundent fermentable carbon source; n + c, limiting nitrogen and nonfermentable carbon source; −, no nutrients.
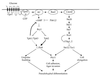
FIG. 2
Filamentous and invasive growth of S. cerevisiae. On the left, invasive growth of haploid and diploid wild-type (wt) and tpk2 mutant yeast colonies is shown. On the right, the typical morphology of yeast pseudohyphal colonies is depicted, and a tpk2/tpk2 mutant with a filamentous growth defect is shown for comparison.
FIG. 3
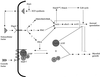
Signal transduction cascades regulating pseudohyphal differentiation in S. cerevisae. Two parallel signaling cascades regulating filamentous growth are depicted: a nutrient-sensing cAMP-PKA pathway, and the MAP kinase cascade. AC, adenylylcyclase.
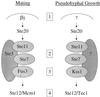
FIG. 4
Signaling specificity during mating and pseudohyphal growth in S. cerevisiae. Two related MAP kinase signaling cascades allow haploid and diploid yeast cells to respond to two different environmental signs, pheromone and nitrogen limitation, and give rise to two completely different developmental fates, mating in haploid cells and filamentous growth in diploid cells. Signaling specificity is achieved by at least four mechanisms. First, pheromone activates a G protein whose βγ subunits activate the MAP kinase cascade, and these components are not expressed in diploid yeast cells and do not regulate filamentous growth. Second, the Ste5 scaffold tethers the components during mating but does not play a role in diploid filamentous growth. Third, the MAP kinase has diverged and specialized: Fus3 to regulate mating and Kss1 to regulate filamentous growth. Finally, Ste12 homodimers or heterodimers with Mcm1 activate pheromone response element-regulated genes in mating or with Tec1 to regulate filamentation response element-driven genes during diploid filamentous growth.
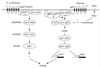
FIG. 5
Signaling pathways regulating mating of S. pombe. Similar to S. cerevisiae diploid pseudohyphal growth, mating in S. pombe is regulated by two parallel signaling pathways. One responds to pheromones and involves a conserved MAP kinase (MAPK) signaling pathway. The second is a nutrient-sensing pathway that, in the presence of abundant nutrients, stimulates cAMP production to repress ste11 expression and inhibit mating. CAP, cyclase-associated protein.
FIG. 6
Role of G proteins in pheromone response in budding and fission yeasts and the human fungal pathogen C. neoformans. The G protein subunits regulating pheromone and nutrient responses in these organisms are depicted. Dashed arrows depict weaker signaling.
FIG. 7
Role of G proteins in filamentous growth and mating in budding and fission yeasts. In both yeasts, two Gα proteins regulate cellular responses to pheromone and nutrients. In both, one functions as a heterotrimeric G protein with a βγ partner, and the other signals in conjunction with Ras. Interestingly, the heterotrimeric G protein is coupled to pheromone receptors in S. cerevisiae but to the nutrient sensor in S. pombe. Importantly, the βγ subunits appear to function with only one of the two Gα subunits in both organisms. MAPK, MAP kinase; AC, adenylylcyclase.
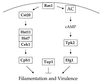
FIG. 8
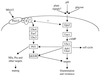
Signaling cascades regulating virulence of C. albicans. Two parallel signaling cascades involving a MAP kinase and a cAMP-PKA signaling pathway regulate filamentation and virulence of this human fungal pathogen. AC, adenylycyclase.
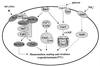
FIG. 9
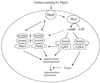
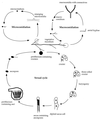
Life cycle of C. neoformans. Mating begins with conjugation tube formation, leading to cell fusion, followed by formation of dikaryotic filaments with fused clamp connections. Terminal basidia form from the tips of mating filaments, where nuclear fusion (karyogamy) occurs. Meiosis and sporulation follow, and long chains of basidiospores are produced, which subsequently germinate into yeast cells. Filamentation independent of a mating partner (haploid fruiting) is observed in MATα strains in response to nitrogen limitation and dessication.
FIG. 10
Signal transduction pathways in C. neoformans. Lightly shaded proteins have been identified in C. neoformans. Darkly shaded proteins are mating type specific. Two of the 24 proteins depicted here have not yet been identified in C. neoformans but are hypothesized from characterization of their roles in other fungi. These are the γ subunit of the pheromone-regulated G protein and the Gpr1 receptor homolog, which are depicted with question marks and dashed symbols. CAM, calmodulin.
FIG. 11
Pheromones stimulate filamentous growth of S. cerevisiae and C. neoformans. (A) Haploid MATa cells of S. cerevisiae are stimulated to invade the agar by pheromone. The disk containing synthetic pheromone was placed on the surface of the lawn of cells, and following growth, the surface of the agar was washed. (B) MATα cells of C. neoformans are stimulated to haploid fruit by confrontation with MATa cells (293). (Panel A was reprinted from reference with permission of the authors and publisher; © 2000, American Association for the Advancement of Science.)
FIG. 12
Life cycle of U. maydis. The life cycle of U. maydis is characterized by a dimorphic switch from a saprophytic yeast form to a pathogenic filamentous growth form. The maintenance of the pathogenic stage is dependent on the presence of the plant host Zea mays.
FIG. 13
Signaling networks in U. maydis. Mating, filamentation, and virulence in the smut fungus U. maydis are regulated by a pheromone response pathway (left) and the cAMP-dependent signaling pathway acting through PKA (Ubc1, Adr1, and Uka1) (right).
FIG. 14
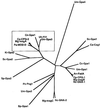
Evolutionary tree showing the relationship between fungal Gα proteins. The branch on the left includes a family of Gα proteins that function in nutrient sensing, some of which are contained within the box. (Adapted from Figure 2 of Loubradou et al. [174] with permission of the authors and publisher; © 1999, Genetics Society of America.) Um, U. maydis; Sc, S. cerevisiae; Ca, C. albicans; An, A. nidulans; Cp, C. parasitica; Mg, M. grisea; Nc, N. crassa; Cn, C. neoformans; Sp, S. pombe; Cc, Coprinus congregatus; Uh, Ustilago hordei; Pa, Podospora anserina; Kl, Kluyveromyces lactis; Pc, Pneumocystis carinii..
FIG. 15
Role of cAMP in fungal morphogenesis. The nutrient-sensing receptor-G protein-cAMP-PKA signaling pathways are depicted. Two related receptors, Gpr1 and git3, play a conserved role in glucose sensing in S. cerevisiae and S. pombe; homologous receptors in C. neoformans and U. maydis remain to be identified. The receptors are coupled to a highly conserved Gα protein (variously called Gpa2, gpa2, Gpa1, and Gpa3), and the C-terminal domain that is known to interact with receptor has >80% sequence identify. The Gα protein is coupled to adenylyl cyclase, and cAMP regulates PKA. Targets of PKA largely remain to be identified but will likely include a variety of transcription factors. Note that cAMP functions to activate or inhibit development depending on the particular organism. AC, adenylyl cyclase.
FIG. 16
Signaling pathways regulating virulence of the rice blast fungus M. grisea. Appressorium formation and virulence of the rice blast fungus M. grisea involve both conserved MAP kinase signaling cascades and the cAMP-PKA signaling pathway.
FIG. 17
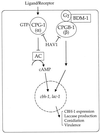
Signaling pathways regulating virulence of the chestnut blight fungus C. parasitica; current model of the G protein signaling pathways that affect virulence and hypovirulence as a result of signal perturbation by HAV in C. parasitica. The Gα protein CPG-1 negatively regulates a putative adenylyl cyclase (AC), whereas the Gβ protein CPGB-1 and BDM-1 may function via a different pathway to regulate gene expression. HAV is hypothesized to function on G proteins or upstream targets.
FIG. 18
Asexual sporulation cycle of A. nidulans. Asexual sporulation of A. nidulans involves two phases: a vegetative growth phase, in which spore germlings give rise to hyphae and the mycelium, and an asexual reproductive phase, in which specialized hyphal cells called foot cells give rise to aerial stalks that produce spore-bearing structures called conidiophores. Stalks produce a vesicle that buds to produce branching cells called sterigmata. Further mitotic divisions of secondary sterigmata called phialides give rise to chains of conidia which germinate on an appropriate medium to complete the cycle.
FIG. 19
Model for the signal transduction pathways regulating asexual sporulation in A. nidulans. Initiation of asexual sporulation in A. nidulans is governed by two parallel antagonistic pathways. Initial mycelial growth is driven by activation of the Gα protein FadA to its GTP-bound form. This pathway represses sporulation until the fungus gains developmental competence. FluG is necessary for the synthesis of an extracellular factor (ECF) that initiates the asexual sporulation pathway by inducing the activities of FlbA, FlbB, FlbC, FlbD, and FlbE, resulting in expression of the brlA, abaA, and wetA genes. Additionally, stimulation of FlbA causes inactivation of FadA, releasing the inhibition of sporulation. The progression of sporulation is also influenced by brlA_-dependent induction of the activities of cell cycle kinases NimX_cdc2 and NimA, promoting morphogenesis from the multinucleate, filamentous form to a uninucleate, budding form.
FIG. 20
Life cycle of N. crassa. In response to environmental conditions, the vegetative mycelium produces conidia (macroconidia and microconidia) and protoperithecia. Macroconidia arise from conidiophores that differentiate from aerial hyphae, and microconidia arise directly from vegetative hyphae. Once a protoperithecium is fertilized with a male organ, sexual development ensues and ascospores are produced.
FIG. 21
Model for signal transduction pathways in N. crassa. A cAMP signaling pathway consisting of G protein α subunits (GNA1 and GNA2), adenylyl cyclase (CR1), and a PKA regulatory subunit (MCB) is involved in conidiation, morphogenesis, mating, and stress tolerance. Ras-, MAP kinase (MAPK)-, and PKA-related kinase-mediated signaling pathways also regulate conidiation. How signaling is coordinated between these pathways remains to be elucidated.
Similar articles
- Conserved cAMP signaling cascades regulate fungal development and virulence.
D'Souza CA, Heitman J. D'Souza CA, et al. FEMS Microbiol Rev. 2001 May;25(3):349-64. doi: 10.1111/j.1574-6976.2001.tb00582.x. FEMS Microbiol Rev. 2001. PMID: 11348689 Review. - Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae.
Pan X, Harashima T, Heitman J. Pan X, et al. Curr Opin Microbiol. 2000 Dec;3(6):567-72. doi: 10.1016/s1369-5274(00)00142-9. Curr Opin Microbiol. 2000. PMID: 11121775 Review. - Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae.
Pan X, Heitman J. Pan X, et al. Mol Cell Biol. 1999 Jul;19(7):4874-87. doi: 10.1128/MCB.19.7.4874. Mol Cell Biol. 1999. PMID: 10373537 Free PMC article. - Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways.
Kronstad J, De Maria AD, Funnell D, Laidlaw RD, Lee N, de Sá MM, Ramesh M. Kronstad J, et al. Arch Microbiol. 1998 Nov;170(6):395-404. doi: 10.1007/s002030050659. Arch Microbiol. 1998. PMID: 9799282 Review. - ras2 Controls morphogenesis, pheromone response, and pathogenicity in the fungal pathogen Ustilago maydis.
Lee N, Kronstad JW. Lee N, et al. Eukaryot Cell. 2002 Dec;1(6):954-66. doi: 10.1128/EC.1.6.954-966.2002. Eukaryot Cell. 2002. PMID: 12477796 Free PMC article.
Cited by
- Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae.
Zhang H, Tang W, Liu K, Huang Q, Zhang X, Yan X, Chen Y, Wang J, Qi Z, Wang Z, Zheng X, Wang P, Zhang Z. Zhang H, et al. PLoS Pathog. 2011 Dec;7(12):e1002450. doi: 10.1371/journal.ppat.1002450. Epub 2011 Dec 29. PLoS Pathog. 2011. PMID: 22241981 Free PMC article. - Paracoccidoides brasiliensis 30 kDa adhesin: identification as a 14-3-3 protein, cloning and subcellular localization in infection models.
da Silva Jde F, de Oliveira HC, Marcos CM, da Silva RA, da Costa TA, Calich VL, Almeida AM, Mendes-Giannini MJ. da Silva Jde F, et al. PLoS One. 2013 Apr 30;8(4):e62533. doi: 10.1371/journal.pone.0062533. Print 2013. PLoS One. 2013. PMID: 23638109 Free PMC article. - Signaling governed by G proteins and cAMP is crucial for growth, secondary metabolism and sexual development in Fusarium fujikuroi.
Studt L, Humpf HU, Tudzynski B. Studt L, et al. PLoS One. 2013;8(2):e58185. doi: 10.1371/journal.pone.0058185. Epub 2013 Feb 28. PLoS One. 2013. PMID: 23469152 Free PMC article. - The phosducin-like protein PhLP1 impacts regulation of glycoside hydrolases and light response in Trichoderma reesei.
Tisch D, Kubicek CP, Schmoll M. Tisch D, et al. BMC Genomics. 2011 Dec 19;12:613. doi: 10.1186/1471-2164-12-613. BMC Genomics. 2011. PMID: 22182583 Free PMC article. - Ras GTPase-activating protein regulation of actin cytoskeleton and hyphal polarity in Aspergillus nidulans.
Harispe L, Portela C, Scazzocchio C, Peñalva MA, Gorfinkiel L. Harispe L, et al. Eukaryot Cell. 2008 Jan;7(1):141-53. doi: 10.1128/EC.00346-07. Epub 2007 Nov 26. Eukaryot Cell. 2008. PMID: 18039943 Free PMC article.
References
- Adams T H, Boylan M T, Timberlake W E. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell. 1988;54:353–362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 AI039115/AI/NIAID NIH HHS/United States
- R01 AI042159/AI/NIAID NIH HHS/United States
- R01 AI42159/AI/NIAID NIH HHS/United States
- R01 AI039115/AI/NIAID NIH HHS/United States
- R01 AI41937/AI/NIAID NIH HHS/United States
- R01 AI39115/AI/NIAID NIH HHS/United States
- P01 AI044975/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases