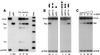Stimulation of homologous recombination through targeted cleavage by chimeric nucleases - PubMed (original) (raw)
Stimulation of homologous recombination through targeted cleavage by chimeric nucleases
M Bibikova et al. Mol Cell Biol. 2001 Jan.
Abstract
Chimeric nucleases that are hybrids between a nonspecific DNA cleavage domain and a zinc finger DNA recognition domain were tested for their ability to find and cleave their target sites in living cells. Both engineered DNA substrates and the nucleases were injected into Xenopus laevis oocyte nuclei, in which DNA cleavage and subsequent homologous recombination were observed. Specific cleavage required two inverted copies of the zinc finger recognition site in close proximity, reflecting the need for dimerization of the cleavage domain. Cleaved DNA molecules were activated for homologous recombination; in optimum conditions, essentially 100% of the substrate recombined, even though the DNA was assembled into chromatin. The original nuclease has an 18-amino-acid linker between the zinc finger and cleavage domains, and this enzyme cleaved in oocytes at paired sites separated by spacers in the range of 6 to 18 bp, with a rather sharp optimum at 8 bp. By shortening the linker, we found that the range of effective site separations could be narrowed significantly. With no intentional linker between the binding and cleavage domains, only binding sites exactly 6 bp apart supported efficient cleavage in oocytes. We also showed that two chimeric enzymes with different binding specificities could collaborate to stimulate recombination when their individual sites were appropriately placed. Because the recognition specificity of zinc fingers can be altered experimentally, this approach holds great promise for inducing targeted recombination in a variety of organisms.
Figures
FIG. 1
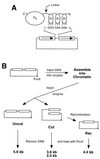
(A) Schematic diagram of a chimeric nuclease and DNA substrate. The nuclease consists of three zinc fingers (Zn) connected to the cleavage domain of _Fok_I (FN) by a flexible peptide linker. The N and C termini of the protein are indicated. Each finger makes contact with three consecutive base pairs in the recognition sequence. In the DNA substrates, the canonical binding site for QQR, 5′-GGG GAA GAA, was inserted, in various numbers and orientations, between the 1.25-kb direct repeats (boxes with arrows) of plasmid pRW4. (B) Scheme for the oocyte injection experiments. The DNA substrate is diagrammed at the top left, and the position of the unique _Pvu_II site is shown. For each sample the DNA was injected into the nuclei of 20 to 40 oocytes; they were incubated for 3 to 4 h to allow chromatin assembly, and then QQR was injected into the nuclei. After various lengths of time, DNA was recovered from the oocytes, digested with _Pvu_II, and analyzed by Southern blot hybridization. This distinguishes DNA molecules not cleaved by QQR (Uncut) from cleaved DNA (Cut) and from cleaved molecules that have undergone homologous recombination (Rec). The locations of the _Pvu_II sites and the sizes of the resulting _Pvu_II fragments are shown.
FIG. 2
Cleavage and recombination in oocytes. (A) Time course of cleavage and recombination in Xenopus oocytes after injection of QQR. Circular pQT10 DNA (0.3 fmol; this corresponds to 0.6 fmol of binding sites) was injected into oocyte nuclei, following the scheme shown in Fig. 1B; 10 nl (30 fmol) of a solution of QQR was delivered to each oocyte, and DNA was recovered at the indicated times after enzyme injection. Recovered DNA was analyzed as described in the legend to Fig. 1B; the positions of the expected fragments (Uncut, Cut, and Rec) are indicated. pHSS6 is a circular plasmid that was included as a recovery control (22). Uninjected pQT10 (Uninj) is shown in the first lane, and uninjected pBR322 (pBR) serves as a marker for the position of recombination products. Stds, linear size standards. The percentage of the recovered DNA that was in the product band (Rec) is indicated below each lane. (B) Effect of recognition site disposition on cleavage and recombination. The number and orientation of sites are indicated by the arrowheads above each lane, with the point designating the A end of the binding site. pQS has a single recognition site; pQT10 has two sites in tail-to-tail inverted orientation; the two sites in pQH10 are in head-to-head orientation; pQD10 has two sites in direct repeat orientation; and pQDD10 has three direct repeats. Each pair of neighboring sites is separated by 10 bp. DNA and enzyme concentrations were as in panel A; incubation was done overnight. (C) Cleavage and recombination in oocytes after injection of various amounts of QQR (3 fmol/nl), as indicated. The DNA substrate was pQT10, and incubation was overnight.
FIG. 3
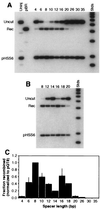
Effect of spacer length on efficiency of cleavage and recombination. All substrates carried two recognition sites in tail-to-tail orientation, separated by the number of base pairs given above the lanes. Labels and markers are as in Fig. 2. Two independent experiments are shown, one (A) covering a broad range of recognition site separations and the other (B) focusing on the range from 12 to 20 bp. About 0.3 fmol of DNA and 70 fmol of QQR were injected in each sample. (C) Histogram summarizing the results of several independent oocyte injection experiments. Recombination yields are plotted against the distance in base pairs between inverted sites. Values were normalized to the recombination fraction measured for pQT8 in each experiment; thin lines represent standard deviations from three or four independent experiments. The values for spacers of 14 and 18 bp are based on a single experiment but were confirmed qualitatively by two additional observations.
FIG. 4
Molecular model of the domains of the chimeric nuclease on DNA. The cleavage domain dimer (ball representation in transparent wheat) sits largely behind the DNA (white) in this view and reaches around the duplex at the top and bottom. The zinc finger domains wind through the major groove and are shown in ribbon representation centered around the cleavage dimer: cyan for binding sites separated by 6 bp and red for a 10-bp separation. The residues that must be connected by the flexible linker are colored green on the zinc finger domains and dark blue on the cleavage domains. In moving from the 6-bp to the 10-bp separation, the attachment sites for the linker have retracted both axially and into the plane of the picture. The distance that the linker must extend to join the binding and cleavage domains has gone from about 20 Å for the 6-bp spacer to >30 Å for the 10-bp case. If the linker cannot reach this distance once the zinc finger domains are bound, the cleavage domain cannot dimerize. This model (52) was produced with the program O, and the figure was generated with MolScript.
FIG. 5
Activity of the L0 linker variant. Two separate experiments showing cleavage and recombination of substrates with various spacer lengths (indicated above each lane) after injection of L0 nuclease. DNA (0.3 fmol) was injected, followed by 5 fmol of enzyme in A and 10 fmol of enzyme in B. Other conditions are as in Fig. 2.
FIG. 6
Summary of the activities of all linker variants in oocytes. Results of experiments like those in Fig. 3 and 6 were quantitated, and the percent recombination is plotted against spacer length (in base pairs) for each enzyme. In all experiments 0.3 fmol of DNA and approximately 10 fmol of nuclease were injected. For L18 (QQR), this is considerably less enzyme than was used in the experiments shown in Fig. 2.
FIG. 7
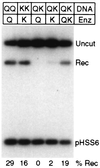
Cleavage and recombination with paired nonidentical sites. The DNA substrates carried two sites for QQR (QQ), two sites for QNK (KK), or one each for QQR and QNK (QK). They were injected into oocytes followed by no enzyme (−), with a single enzyme (Q or K), or with a mixture of the two enzymes (QK). In each case, 0.3 fmol of DNA and 15 fmol of each enzyme were injected.
Similar articles
- Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains.
Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Smith J, et al. Nucleic Acids Res. 2000 Sep 1;28(17):3361-9. doi: 10.1093/nar/28.17.3361. Nucleic Acids Res. 2000. PMID: 10954606 Free PMC article. - Chimeric nucleases stimulate gene targeting in human cells.
Porteus MH, Baltimore D. Porteus MH, et al. Science. 2003 May 2;300(5620):763. doi: 10.1126/science.1078395. Science. 2003. PMID: 12730593 No abstract available. - High-frequency homologous recombination in plants mediated by zinc-finger nucleases.
Wright DA, Townsend JA, Winfrey RJ Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. Wright DA, et al. Plant J. 2005 Nov;44(4):693-705. doi: 10.1111/j.1365-313X.2005.02551.x. Plant J. 2005. PMID: 16262717 - Chimeric restriction enzymes: what is next?
Chandrasegaran S, Smith J. Chandrasegaran S, et al. Biol Chem. 1999 Jul-Aug;380(7-8):841-8. doi: 10.1515/BC.1999.103. Biol Chem. 1999. PMID: 10494832 Free PMC article. Review. - [Zinc finger nucleases and their application].
Deng SS, Wang YZ, Ma D. Deng SS, et al. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010 Apr;27(2):162-5. doi: 10.3760/cma.j.issn.1003-9406.2010.02.010. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010. PMID: 20376797 Review. Chinese.
Cited by
- Engineered PsCas9 enables therapeutic genome editing in mouse liver with lipid nanoparticles.
Degtev D, Bravo J, Emmanouilidi A, Zdravković A, Choong OK, Liz Touza J, Selfjord N, Weisheit I, Francescatto M, Akcakaya P, Porritt M, Maresca M, Taylor D, Sienski G. Degtev D, et al. Nat Commun. 2024 Nov 7;15(1):9173. doi: 10.1038/s41467-024-53418-8. Nat Commun. 2024. PMID: 39511150 Free PMC article. - Genome editing in Sub-Saharan Africa: a game-changing strategy for climate change mitigation and sustainable agriculture.
Amoah P, Oumarou Mahamane AR, Byiringiro MH, Mahula NJ, Manneh N, Oluwasegun YR, Assfaw AT, Mukiti HM, Garba AD, Chiemeke FK, Bernard Ojuederie O, Olasanmi B. Amoah P, et al. GM Crops Food. 2024 Dec 31;15(1):279-302. doi: 10.1080/21645698.2024.2411767. Epub 2024 Oct 31. GM Crops Food. 2024. PMID: 39481911 Free PMC article. Review. - Identification and Validation of New DNA-PKcs Inhibitors through High-Throughput Virtual Screening and Experimental Verification.
Dai L, Yu P, Fan H, Xia W, Zhao Y, Zhang P, Zhang JZH, Zhang H, Chen Y. Dai L, et al. Int J Mol Sci. 2024 Jul 22;25(14):7982. doi: 10.3390/ijms25147982. Int J Mol Sci. 2024. PMID: 39063224 Free PMC article. - Increasing Knockin Efficiency in Mouse Zygotes by Transient Hypothermia.
Bouchareb A, Biggs D, Alghadban S, Preece C, Davies B. Bouchareb A, et al. CRISPR J. 2024 Apr;7(2):111-119. doi: 10.1089/crispr.2023.0077. CRISPR J. 2024. PMID: 38635329 Free PMC article. - Gene editing in small and large animals for translational medicine: a review.
Mariano CG, de Oliveira VC, Ambrósio CE. Mariano CG, et al. Anim Reprod. 2024 Apr 12;21(1):e20230089. doi: 10.1590/1984-3143-AR2023-0089. eCollection 2024. Anim Reprod. 2024. PMID: 38628493 Free PMC article. Review.
References
- Capecchi M R. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. - PubMed
- Carroll D. DNA recombination and repair in Xenopus oocytes and eggs: substrate design, direct microinjection, and extract preparation. In: Richter J D, editor. A comparative methods approach to the study of oocytes and embryos. New York, N.Y: Oxford University Press; 1999. pp. 173–195.
- Carroll D. Homologous genetic recombination in Xenopus: mechanism and implications for gene manipulation. Prog Nucleic Acid Res Mol Biol. 1996;54:101–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM50739/GM/NIGMS NIH HHS/United States
- GM58504/GM/NIGMS NIH HHS/United States
- R01 GM058504/GM/NIGMS NIH HHS/United States
- R01 GM053923/GM/NIGMS NIH HHS/United States
- GM53923/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials