Statistical methods for detecting molecular adaptation - PubMed (original) (raw)
Statistical methods for detecting molecular adaptation
Z Yang et al. Trends Ecol Evol. 2000.
Abstract
The past few years have seen the development of powerful statistical methods for detecting adaptive molecular evolution. These methods compare synonymous and nonsynonymous substitution rates in protein-coding genes, and regard a nonsynonymous rate elevated above the synonymous rate as evidence for darwinian selection. Numerous cases of molecular adaptation are being identified in various systems from viruses to humans. Although previous analyses averaging rates over sites and time have little power, recent methods designed to detect positive selection at individual sites and lineages have been successful. Here, we summarize recent statistical methods for detecting molecular adaptation, and discuss their limitations and possible improvements.
Figures
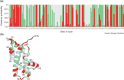
Fig. 1
The identification of sites under positive selection from the sperm lysin genes of 25 abalone species. (a) Posterior probabilities for site classes with different ω ratios along the sequence. The lysin sequence of the red abalone (Haliotis rufescens) is shown below the x-axis. ML estimates under Model M3 (discrete) suggest three site classes with the ω ratios at ω0=0.085 (grey), ω1=0.911 (green) and ω2=3.065 (red), and with proportions _p_0=0.329, _ρ_1=0.402 and _ρ_2=0.269. These proportions are the prior probabilities (Box 1) that any site belongs to the three classes. The data (codon configurations in different species) at a site alter the prior probabilities dramatically, and thus the posterior probabilities might be different from the prior probabilities. For example, the posterior probabilities for Site 1 are 0.944, 0.056 and 0.000, and thus this site is likely to be under strong purifying selection. The posterior probabilities for Site 4 are 0.000, 0.000 and 1.000, and thus this site is almost certainly under diversifying selection. (b) Lysin crystal structure from the red abalone (Protein Data Bank file 1LIS), with sites coloured according to their most likely class inferred in (a). The structure starts from amino acid four (His) at the N-terminus, because the first three amino acids are unresolved. The five α-helices are indicated: α1 from amino acids 13 to 38, α2 from 44 to 74, α3 from 82 to 95, α4 from 99 to 107 and α5 from 116 to 123. Note that sites potentially under positive selection (red) are scattered all over the primary sequence but tend to cluster around the top and bottom of the crystal structure. Reproduced, with permission, from.
Similar articles
- Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages.
Yang Z, Nielsen R. Yang Z, et al. Mol Biol Evol. 2002 Jun;19(6):908-17. doi: 10.1093/oxfordjournals.molbev.a004148. Mol Biol Evol. 2002. PMID: 12032247 - Maximum likelihood methods for detecting adaptive evolution after gene duplication.
Bielawski JP, Yang Z. Bielawski JP, et al. J Struct Funct Genomics. 2003;3(1-4):201-12. J Struct Funct Genomics. 2003. PMID: 12836699 Review. - Inference of selection from multiple species alignments.
Yang Z. Yang Z. Curr Opin Genet Dev. 2002 Dec;12(6):688-94. doi: 10.1016/s0959-437x(02)00348-9. Curr Opin Genet Dev. 2002. PMID: 12433583 Review. - Molecular evolution of nuclear genes in Cupressacea, a group of conifer trees.
Kusumi J, Tsumura Y, Yoshimaru H, Tachida H. Kusumi J, et al. Mol Biol Evol. 2002 May;19(5):736-47. doi: 10.1093/oxfordjournals.molbev.a004132. Mol Biol Evol. 2002. PMID: 11961107 - Not so different after all: a comparison of methods for detecting amino acid sites under selection.
Kosakovsky Pond SL, Frost SD. Kosakovsky Pond SL, et al. Mol Biol Evol. 2005 May;22(5):1208-22. doi: 10.1093/molbev/msi105. Epub 2005 Feb 9. Mol Biol Evol. 2005. PMID: 15703242
Cited by
- Comparison of the chloroplast genomics of nine endangered Habenaria species and phylogenetic analysis.
Zhang J, Zhou D, Chen W, Lin P, Zhao S, Wang M, Wang H, Shi S, Mehmood F, Ye X, Meng J, Zhuang W. Zhang J, et al. BMC Plant Biol. 2024 Nov 5;24(1):1046. doi: 10.1186/s12870-024-05766-2. BMC Plant Biol. 2024. PMID: 39497089 Free PMC article. - Intra- and inter-subtype HIV diversity between 1994 and 2018 in southern Uganda: a longitudinal population-based study.
Kim S, Kigozi G, Martin MA, Galiwango RM, Quinn TC, Redd AD, Ssekubugu R, Bonsall D, Ssemwanga D, Rambaut A, Herbeck JT, Reynolds SJ, Foley B, Abeler-Dörner L, Fraser C, Ratmann O, Kagaayi J, Laeyendecker O, Grabowski MK. Kim S, et al. Virus Evol. 2024 Aug 24;10(1):veae065. doi: 10.1093/ve/veae065. eCollection 2024. Virus Evol. 2024. PMID: 39399152 Free PMC article. - Taxonomic Distribution, Phylogenetic Relationship, and Domain Conservation of CRISPR-Associated Cas Proteins.
Ranasinghe W, Gillette D, Ho A, Cho H, Choudhary M. Ranasinghe W, et al. Bioinform Biol Insights. 2024 Oct 5;18:11779322241274961. doi: 10.1177/11779322241274961. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 39397878 Free PMC article. - Characteristics and Comparative Analysis of Six Mitogenomes of Genus Kiefferulus Goetghebuer, 1922 (Diptera: Chironomidae).
Zhang D, Jin WD, Xu HF, Li XB, Jiang YW, Li DQ, Lin XL. Zhang D, et al. Insects. 2024 Aug 28;15(9):646. doi: 10.3390/insects15090646. Insects. 2024. PMID: 39336614 Free PMC article. - Signals of positive selection in genomes of palearctic Myotis-bats coexisting with a fungal pathogen.
Twort VG, Laine VN, Field KA, Whiting-Fawcett F, Ito F, Reiman M, Bartonicka T, Fritze M, Ilyukha VA, Belkin VV, Khizhkin EA, Reeder DM, Fukui D, Jiang TL, Lilley TM. Twort VG, et al. BMC Genomics. 2024 Sep 3;25(1):828. doi: 10.1186/s12864-024-10722-3. BMC Genomics. 2024. PMID: 39227786 Free PMC article.
References
- Lewontin R.C. Adaptation. Sci. Am. 1979;239:156–169. - PubMed
- Kimura M. The Neutral Theory of Molecular Evolution. University Press; Cambridge: 1983.
- Endo T. Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 1996;13:685–690. - PubMed
- Hughes A.L. Adaptive Evolution of Genes and Genomes. University Press; Oxford: 1999.
- Miyata T., Yasunaga T. Molecular evolution of mRNA: a method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its applications. J. Mol. Evol. 1980;16:23–36. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous