Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens - PubMed (original) (raw)
Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens
J R Fitzgerald et al. J Bacteriol. 2001 Jan.
Erratum in
- J Bacteriol 2001 Mar 15;166(6):following 4259
Abstract
Previous studies have demonstrated that a proportion of Staphylococcus aureus isolates from bovine mastitis coproduce toxic shock syndrome toxin (TSST) and staphylococcal enterotoxin C (SEC). In this study, molecular genetic analysis of one such strain, RF122, revealed the presence of a 15,891-bp putative pathogenicity island (SaPIbov) encoding the genes for TSST (tst), the SEC bovine variant (sec-bovine), and a gene (sel) which encodes an enterotoxin-like protein. The island contains 21 open reading frames specifying hypothetical proteins longer than 60 amino acids including an integrase-like gene. The element is bordered by 74-bp direct repeats at the left and right junctions, and the integration site lies adjacent to the 3' end of the GMP synthase gene (gmps) in the S. aureus chromosome. SaPIbov contains a central region of sequence identity with the previously characterized tst pathogenicity island SaPI1 (J. A. Lindsay et al., Mol. Microbiol. 29:527-543, 1998). A closely related strain, RF120, of the same multilocus enzyme electrophoretic type, random amplified polymorphic DNA type, and ribotype, does not contain the island, implying that the element is mobile and that a recent insertion/deletion event has taken place. TSST and TSST/SEC-deficient mutants of S. aureus strain RF122 were constructed by allele replacement. In vitro bovine Vbeta-specific lymphocyte expansion analysis by culture supernatants of wild-type strains and of tst and sec-bovine allele replacement mutants revealed that TSST stimulates BTB13-specific T cells whereas SEC-bovine stimulates BTB93-specific T cells. This suggests that the presence of SaPIbov may contribute to modulation of the bovine immune response.
Figures
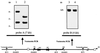
FIG. 1
Mapping SaPIbov by PCR and Southern hybridization. Vectorette PCR was performed on DNA flanking the 6.5-kb _Hin_dIII fragment harboring tst and sec. DNA was cleaved with _Bcl_I, ligated with the Vectorette cassette, and subjected to PCR with outward-directed primer VL or VR and a primer specific for the Vectorette unit attached to the end of each fragment. These PCR products provided probes A and B, which were used to analyze genomic DNA of strain RF122 tst sec (lanes 1 and 3) and wild-type strain RF120 (lanes 2 and 4). Primers VR and JR1 were used to amplify the 9-kb right junction fragment. The left and right junctions of SaPIbov are denoted by open boxes.
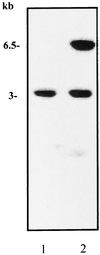
FIG. 2
Southern blot analysis of _Hin_dIII-digested genomic DNA from strain RF120 (lane 1) and strain RF122 tst sec (lane 2) using pJRF101 containing the 6.5-kb _Hin_dIII fragment as a probe. The probe hybridized to a 6.5-kbp fragment of RF122 and also, unexpectedly, to a second 3-kb fragment of both strains, suggesting that the 6.5-kb tst/sec fragment contains homologous sequences elsewhere in the genome.
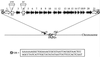
FIG. 3
The 15,891-bp SaPIbov. Black arrows indicate ORFs of unknown function. White arrows indicate genes referred to in the text: sec, sel, int, tst, and gmps. The hatched boxes indicate positions of the direct repeats (DR), the sequence of which is given. Open boxes represent the erythromycin (erm) and tetracycline (tet) cassettes used to construct the TSST− and TSST− SEC− mutants. Primer juncf3 was used in Vectorette PCR to amplify DNA containing the SaPIbov integration site in strain RF120 after digestion with _Alu_I.
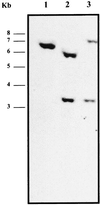
FIG. 4
Southern blot hybridization analysis of tst and tst sec mutants. Genomic DNA from wild-type strain RF122 (lane 1), mutant RF122-1 tst (lane 2), and double mutant RF122-2 tst sec (lane 3) was cleaved with _Hin_dIII, separated by agarose electrophoresis, and transferred to a nylon membrane. It was hybridized with a probe specific for tst covering the site of insertion of the tet marker.
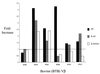
FIG. 5
Representative analysis of boVβ expansion in bovine lymphocyte cultures. Bovine lymphocyte cultures were treated with protein preparations recovered from cultures of S. aureus RF122 (wt [wild-type]), S. aureus RF122-1 (Δ_tst_), and S. aureus RF122-2 (Δ_tst/sec_). Fold increase reflects the fold difference in measured PDU for each boVβ. This value was determined by quantifying reaction products (in PDU) generated from treated lymphocytes to those from an identical but untreated cell culture. All boVβ PDU values were normalized by quantifying bovine lymphocyte TCR Cα levels.
Similar articles
- Occurrence and clonal relatedness of sec/tst-gene positive Staphylococcus aureus isolates of quartermilk samples of cows suffering from mastitis.
Zschöck M, Risse K, Sommerhäuser J. Zschöck M, et al. Lett Appl Microbiol. 2004;38(6):493-8. doi: 10.1111/j.1472-765X.2004.01519.x. Lett Appl Microbiol. 2004. PMID: 15130145 - Genetic diversity and virulence characteristics of Staphylococcus aureus isolates from cases of bovine mastitis.
Vaughn JM, Abdi RD, Gillespie BE, Kerro Dego O. Vaughn JM, et al. Microb Pathog. 2020 Jul;144:104171. doi: 10.1016/j.micpath.2020.104171. Epub 2020 Mar 26. Microb Pathog. 2020. PMID: 32224210 - Genotyping of long term persistent Staphylococcus aureus in bovine subclinical mastitis.
Rossi BF, Bonsaglia ECR, Castilho IG, Dantas STA, Salina A, Langoni H, Pantoja JCF, Budri PE, Fitzgerald-Hughes D, Júnior AF, Rall VLM. Rossi BF, et al. Microb Pathog. 2019 Jul;132:45-50. doi: 10.1016/j.micpath.2019.04.031. Epub 2019 Apr 20. Microb Pathog. 2019. PMID: 31015015 - Detection of superantigenic toxin genes in Staphylococcus aureus strains from subclinical bovine mastitis.
Günaydın B, Aslantaş Ö, Demir C. Günaydın B, et al. Trop Anim Health Prod. 2011 Dec;43(8):1633-7. doi: 10.1007/s11250-011-9882-5. Epub 2011 Jun 4. Trop Anim Health Prod. 2011. PMID: 21643668 - Diversity in Staphylococcus aureus enterotoxins.
Thomas D, Chou S, Dauwalder O, Lina G. Thomas D, et al. Chem Immunol Allergy. 2007;93:24-41. doi: 10.1159/000100856. Chem Immunol Allergy. 2007. PMID: 17369698 Review.
Cited by
- Detection of the enterotoxigenic genes (sei,sej) in Staphylococcus aureus isolates from bovine mastitis milk in the West Azerbaijan of Iran.
Ahmady M, Kazemi S. Ahmady M, et al. Comp Clin Path. 2013 Jul;22(4):649-654. doi: 10.1007/s00580-012-1460-3. Epub 2012 Mar 15. Comp Clin Path. 2013. PMID: 23864850 Free PMC article. - Silence as a way of niche adaptation: mecC-MRSA with variations in the accessory gene regulator (agr) functionality express kaleidoscopic phenotypes.
Huber C, Stamm I, Ziebuhr W, Marincola G, Bischoff M, Strommenger B, Jaschkowitz G, Marciniak T, Cuny C, Witte W, Doellinger J, Schaudinn C, Thürmer A, Epping L, Semmler T, Lübke-Becker A, Wieler LH, Walther B. Huber C, et al. Sci Rep. 2020 Sep 8;10(1):14787. doi: 10.1038/s41598-020-71640-4. Sci Rep. 2020. PMID: 32901059 Free PMC article. - Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic.
Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM. Fitzgerald JR, et al. Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8821-6. doi: 10.1073/pnas.161098098. Epub 2001 Jul 10. Proc Natl Acad Sci U S A. 2001. PMID: 11447287 Free PMC article. - Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: evidence for horizontal transfer of virulence genes.
Moore PC, Lindsay JA. Moore PC, et al. J Clin Microbiol. 2001 Aug;39(8):2760-7. doi: 10.1128/JCM.39.8.2760-2767.2001. J Clin Microbiol. 2001. PMID: 11473989 Free PMC article. - Biological properties of staphylococcal enterotoxin-like toxin type R.
Omoe K, Imanishi K, Hu DL, Kato H, Takahashi-Omoe H, Nakane A, Uchiyama T, Shinagawa K. Omoe K, et al. Infect Immun. 2004 Jun;72(6):3664-7. doi: 10.1128/IAI.72.6.3664-3667.2004. Infect Immun. 2004. PMID: 15155681 Free PMC article.
References
- Augustin J, Gotz F. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett. 1990;66:203–208. - PubMed
- Bayles K W, Brunskill E W, Iandolo J J, Hruska L L, Huan S, Patee P A, Smiley B K, Yasbin R E. A genetic and molecular characterization of the recA gene from Staphylococcus aureus. Gene. 1994;147:13–20. - PubMed
- Betley M J, Mekalanos J J. Staphylococcal enterotoxin A is encoded by phage. Science. 1985;229:185–187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources