Srb7p is a physical and physiological target of Tup1p - PubMed (original) (raw)
Srb7p is a physical and physiological target of Tup1p
A Gromöller et al. EMBO J. 2000.
Abstract
The holoenzyme of transcription integrates the positive and negative signals from the promoters of eukaryotic genes. We demonstrate that the essential holoenzyme component Srb7p is a physiologically relevant target of the global repressor Tup1p in Saccharomyces cerevisiae. Tup1p binds Srb7p in vivo and in vitro, and all genes tested that are repressed by Tup1p are derepressed when wild-type Srb7p is replaced by a mutant derivative of Srb7p that is no longer capable of interacting with Tup1p. Therefore, Srb7p is the first holoenzyme component essential for repression by Tup1p for which a physical interaction with Tup1p has been demonstrated. Furthermore, we find that Srb7p also binds Med6p and that this interaction is necessary for full transcriptional activation by different activators. Our finding that Med6p and Tup1p compete for the interaction with Srb7p suggests a model for Tup1p-mediated repression.
Figures
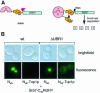
Fig. 1. Tup1p interacts with Srb7p in vivo. (A) The split-ubiquitin system. A fusion containing a protein Y, Cub and the RGFP reporteris not subject to proteolysis by the Ubps. Co-expression of a fusion containing Nub and a protein X capable of interacting with protein Y leads to the formation of a native-like ubiquitin, to subsequent cleavage by the Ubps and to degradation of the RGFP reporter by the enzymes of the N-end rule. (B) The interaction between Tup1p and Srb7p leads to the destruction of the nuclear localized GFP signal in wild-type cells (left), or to the delocalization in cells without a functional N-end rule pathway (ΔUBR1, right). Cells expressing the depicted fusions were grown in liquid medium, spotted onto slides and analysed with the help of a fluorescent microscope (Leitz).
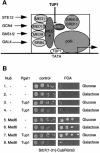
Fig. 2. The N-terminus of Srb7p is necessary and sufficient for the interaction with Tup1p. (A) Tup1p interacts with Srb7p in vivo. Serial dilutions of cells expressing the depicted fusions were spotted onto control plates selecting for the presence of these fusions, onto the same plates lacking uracil and onto plates also containing FOA. Interactions are revealed by the lack of growth on the plates without uracil and by growth on the plates containing FOA. (B) Tup1p interacts with Srb7p in vitro. GST-pulldown assays with the depicted proteins purified from E.coli were loaded onto an SDS gel, and HA-Tup1p was detected in a western blot with the help of an α-HA antibody.
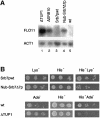
Fig. 3. Srb7p is a physiological target of Tup1p. (A) Disruption of the Tup1p–Srb7p interaction leads to derepression of FLO11. Total RNA was isolated from JD53ΔTUP1 (ΔTUP1), JD53ΔSRB10 (ΔSRB10), JD53ΔSRB7 + Srb7p (Srb7pwt), JD53ΔSRB7 + Nub–Srb7Δ7p (Nub–Srb7Δ7p) and JD53 (wt), and the northern blot was probed with probes derived from the FLO11 and ACT1 genes. (B) Disruption of the Tup1p–Srb7p interaction leads to a decreased mating efficiency. JD53ΔSRB7 + Srb7p (Srb7pwt) and JD53ΔSRB7 + Nub–Srb7Δ7p (Nub–Srb7Δ7p), which are prototrophic for lysine, as well as JD53 (wt) and JD53ΔTUP1 (ΔTUP1), which are prototrophic for adenine, were mated with JD52 + HIS3, which is prototrophic for histidine. Serial dilutions of the crosses were spotted onto the indicated plates and only diploids were able to grow on the double selection plates.
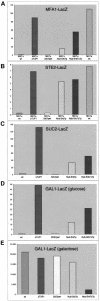
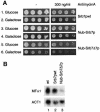
Fig. 4. Srb7p affects transcriptional repression and activation. (A) An MFA1-LacZ fusion was integrated into the MFA1 locus of the strains JD53 (MATα; wt), JD53ΔTUP1 (MATα; ΔTUP1), JD53ΔSRB7 + Srb7p (MATα; Srb7pwt), JD53ΔSRB7 + Nub–Srb7p (MATα; Nub–Srb7p), JD53ΔSRB7 + Nub–Srb7Δ7p (MATα; Nub–Srb7Δ7p) and JD52 (MATa; wt). The cells were grown in liquid culture and β-galactosidase activity was determined. (B) A STE2-LacZ fusion was integrated into the URA3 locus of the same strains as in (A). The cells were grown in liquid culture and β-galactosidase activity was determined. (C) A SUC2-LacZ fusion was integrated into the URA3 locus of the strains described in (A), with the exception of JD52. The cells were grown in liquid culture and β-galactosidase activity was determined. (D) A GAL1-LacZ fusion was integrated into the GAL1 locus of the strains described in (A), with the exception of JD52. The cells were grown on YPDA plates, washed off the plates and β-galactosidase activity was determined. (E) The strains from (D) were grown in liquid medium with 2% galactose, and β-galactosidase activity was determined. All experiments were performed in triplicate and the standard deviation was <20%.
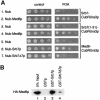
Fig. 5. Med6p interacts with Srb7p in vivo and in vitro. (A) Med6p interacts with Srb7p in vivo. Serial dilutions of cells expressing the depicted fusions were spotted onto control plates selecting for the presence of these fusions and onto plates also containing FOA. Interactions are revealed by growth on the FOA plates. (B) Med6p interacts with Srb7p in vitro. GST-pulldown assays with the depicted proteins were loaded onto an SDS gel and HA-Med6p was detected in a western blot with the help of an α-HA antibody.
Fig. 6. Srb7p is a physiological target of Med6p. (A) Disruption of the Med6p–Srb7p interaction results in a _GAL_– phenotype on plates containing the respiration inhibitor antimycin A. Serial dilutions of the strains JD53ΔSRB7 + Srb7p (Srb7pwt), JD53ΔSRB7 + Nub–Srb7p (Nub–Srb7p) and JD53ΔSRB7 + Nub–Srb7Δ7p (Nub–Srb7Δ7p) were spotted onto plates containing the depicted medium. (B) Disruption of the Med6p–Srb7p interaction leads to a reduction in the transcriptional activation of MFα1. Total RNA was isolated from JD53 (wt), JD53ΔSRB7 + Srb7p (Srb7pwt) and JD53ΔSRB7 + Nub–Srb7Δ7p (Nub–Srb7Δ7p), and the northern blot was probed with probes derived from the MFα1 and ACT1 genes.
Fig. 7. Tup1p works by blocking the access of Med6p to Srb7p. (A) A model for Tup1p action. The interaction between Med6p and Srb7p is needed to transmit positive signals emanating from transcriptional activators to the polymerase, and Tup1p interferes with this interaction. (B) Overexpression of Tup1p inhibits interaction between Med6p and Srb7p. Cells expressing Nub or Nub–Med6p together with the SRB7(1–31)–Cub-RUra3p fusion were transformed with a plasmid expressing Tup1p under the control of the GAL1 promoter, or the empty expression vector. Serial dilutions of the transformants were spotted onto plates containing the depicted medium. The presence of the interaction is revealed by the growth on the FOA plate.
Fig. 8. Sequence alignment of Srb7 proteins from D.melanogaster (DmSrb7p), H.sapiens (HsSrb7p) and S.pombe (SpSrb7p) to Srb7p of S.cerevisiae (ScSrb7p). Residues identical to the consensus sequence shown on top are shaded black. The sequences were derived from the DDBJ/EMBL/GenBank and the alignment was made with the program MegAlign.
Similar articles
- Activator Gcn4p and Cyc8p/Tup1p are interdependent for promoter occupancy at ARG1 in vivo.
Kim SJ, Swanson MJ, Qiu H, Govind CK, Hinnebusch AG. Kim SJ, et al. Mol Cell Biol. 2005 Dec;25(24):11171-83. doi: 10.1128/MCB.25.24.11171-11183.2005. Mol Cell Biol. 2005. PMID: 16314536 Free PMC article. - Genome-wide expression analysis of a Saccharomyces cerevisiae strain deleted for the Tup1p-interacting protein Cdc73p.
Kerkmann K, Lehming N. Kerkmann K, et al. Curr Genet. 2001 Jul;39(5-6):284-90. doi: 10.1007/s002940100229. Curr Genet. 2001. PMID: 11525400 - Srb7p is essential for the activation of a subset of genes.
Gromöller A, Lehming N. Gromöller A, et al. FEBS Lett. 2000 Oct 27;484(1):48-54. doi: 10.1016/s0014-5793(00)02123-2. FEBS Lett. 2000. PMID: 11056220 - The cyclin in the RNA polymerase holoenzyme is a target for the transcriptional repressor Tup1p in Saccharomyces cerevisiae.
Schüller J, Lehming N. Schüller J, et al. J Mol Microbiol Biotechnol. 2003;5(4):199-205. doi: 10.1159/000071071. J Mol Microbiol Biotechnol. 2003. PMID: 12867743 Review. - Mediator of transcriptional regulation.
Björklund S, Kim YJ. Björklund S, et al. Trends Biochem Sci. 1996 Sep;21(9):335-7. doi: 10.1016/s0968-0004(96)10051-7. Trends Biochem Sci. 1996. PMID: 8870496 Review.
Cited by
- The LAMMER kinase homolog, Lkh1, regulates Tup transcriptional repressors through phosphorylation in Schizosaccharomyces pombe.
Kang WH, Park YH, Park HM. Kang WH, et al. J Biol Chem. 2010 Apr 30;285(18):13797-806. doi: 10.1074/jbc.M110.113555. Epub 2010 Mar 3. J Biol Chem. 2010. PMID: 20200159 Free PMC article. - Functional studies of the yeast med5, med15 and med16 mediator tail subunits.
Larsson M, Uvell H, Sandström J, Rydén P, Selth LA, Björklund S. Larsson M, et al. PLoS One. 2013 Aug 22;8(8):e73137. doi: 10.1371/journal.pone.0073137. eCollection 2013. PLoS One. 2013. PMID: 23991176 Free PMC article. - Characterization of mutations that are synthetic lethal with pol3-13, a mutated allele of DNA polymerase delta in Saccharomyces cerevisiae.
Chanet R, Heude M. Chanet R, et al. Curr Genet. 2003 Aug;43(5):337-50. doi: 10.1007/s00294-003-0407-2. Epub 2003 May 21. Curr Genet. 2003. PMID: 12759774 - The TATA-binding protein is not an essential target of the transcriptional activators Gal4p and Gcn4p in Saccharomyces cerevisiae.
Bongards C, Chew BS, Lehming N. Bongards C, et al. Biochem J. 2003 Feb 15;370(Pt 1):141-7. doi: 10.1042/BJ20021548. Biochem J. 2003. PMID: 12423206 Free PMC article. - Cyc8p and Tup1p transcription regulators antagonistically regulate Flo11p expression and complexity of yeast colony biofilms.
Nguyen PV, Hlaváček O, Maršíková J, Váchová L, Palková Z. Nguyen PV, et al. PLoS Genet. 2018 Jul 2;14(7):e1007495. doi: 10.1371/journal.pgen.1007495. eCollection 2018 Jul. PLoS Genet. 2018. PMID: 29965985 Free PMC article.
References
- Ausubel F., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1998) Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York, NY.
- DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. - PubMed
- Dohmen R.J., Stappen,R., McGrath,J.P., Forrova,H., Kolarov,J., Goffeau,A. and Varshavsky,A. (1995) An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem., 270, 18099–18109. - PubMed
- Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases