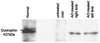Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse - PubMed (original) (raw)
Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse
C J Mann et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Duchenne muscular dystrophy (DMD) is a severe muscle wasting disease arising from defects in the dystrophin gene, typically nonsense or frameshift mutations, that preclude the synthesis of a functional protein. A milder, allelic version of the disease, Becker muscular dystrophy, generally arises from in-frame deletions that allow synthesis of a shorter but still semifunctional protein. Therapies to introduce functional dystrophin into dystrophic tissue through either cell or gene replacement have not been successful to date. We report an alternative approach where 2'-O-methyl antisense oligoribonucleotides have been used to modify processing of the dystrophin pre-mRNA in the mdx mouse model of DMD. By targeting 2'-O-methyl antisense oligoribonucleotides to block motifs involved in normal dystrophin pre-mRNA splicing, we induced excision of exon 23, and the mdx nonsense mutation, without disrupting the reading frame. Exon 23 skipping was first optimized in vitro in transfected H-2K(b)-tsA58 mdx myoblasts and then induced in vivo. Immunohistochemical staining demonstrated the synthesis and correct subsarcolemmal localization of dystrophin and gamma-sarcoglycan in the mdx mouse after intramuscular delivery of antisense oligoribonucleotide:liposome complexes. This approach should reduce the severity of DMD by allowing a dystrophic gene transcript to be modified, such that it can be translated into a Becker-dystrophin-like protein.
Figures
Figure 1
Sequences and relative binding sites of AOs. The sequence of exon 23 of mouse dystrophin is indicated by capitals and a shaded box, whereas the intronic sequences neighboring the exon are indicated by lowercase and a plain box. The mdx mouse has a nonsense mutation at nucleotide 3185 causing premature termination of translation in this exon. The sequence and orientation of all the AOs used in this study are indicated, except for AO-random (5′-CCAGAUCGGA CGACGUCAGG ACAAC-3′), which was designed not to hybridize to any known sequence as validated by a
blast
database search. The fluorescein (FITC) group of AO 5′SS-FITC was attached to the 5′ end of the sequence as indicated.
Figure 2
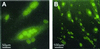
Efficient nuclear uptake of liposome-complexed fluorescein-labeled AO by _H-2K_b-tsA58 mdx cells. Cultured _H-2K_b-tsA58 mdx myotubes were assessed for uptake of fluorescence after exposure to complexes of Lipofectin and AO 5′SS-FITC (2:1 ratio). Nuclear fluorescence was observed in ≈100% of the myotubes 3 h after transfection, with some pinpoint foci of fluorescence located in the cytoplasm and at the cell surface (B). Many of the transfected myotubes displayed multiple fluorescent nuclei (A).
Figure 3
AO-induced exon skipping in_H-2K_b-tsA58 mdx myoblasts. Total RNA was extracted from treated and untreated_H-2K_b-tsA58 mdx cells and amplified by nested RT-PCR using primers annealing to exons 20 and 26. Transfections were carried out as described in the text with the indicated AOs. The 901-bp full-length transcript was detected in all samples except the PCR negative (-ve) control. Lane M, size markers. A shorter product of 688 bp (arrow), corresponding to the removal of exon 23, was amplified from cell extracts transfected with AO 5′SS-FITC and AO 5′SS-25.
Figure 4
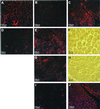
Demonstration of antisense-induced dystrophin synthesis and restoration of γ-sarcoglycan in C57BL mdx mouse muscle. Immunostaining of normal C57BL mouse muscle (A) and untreated C57BL mdx muscle (B) with antibody DYS2 revealed sarcolemmal staining of dystrophin and a complete absence of the protein, respectively. However, sarcolemmal dystrophin staining was observed in a broad spread of_mdx_ muscle fibers 1 week after injection of Lipofectin–AO 5′SS-25 (2:1 ratio) complexes into the left quadriceps-femoris complex (C). At higher magnification, dystrophin staining was revealed to be discontinuous, reminiscent of BMD staining, in both the left (E) and right (G) legs. Phase-contrast light microscopy of the same region (F and H, respectively) shows the green dye used to locate the site of the injection. No positive dystrophin staining was observed in an age-matched mdx control muscle when stained on the same slide as the treated sections and photographed under identical conditions (D). Serial sections of treated muscle stained with antibodies against γ-sarcoglycan revealed restoration of γ-sarcoglycan to the sarcolemma of muscle fibers that also stained positively for dystrophin (J). No staining was observed in the same muscle of an age-matched mdx control section also stained for γ-sarcoglycan (I).
Figure 5
Synthesis of near full-length dystrophin in muscle of_mdx_ mice treated with Lipofectin–AO 5′SS-25 complexes. Western blotting of total protein extracted from the quadriceps-femoris complex of mice treated with Lipofectin–AO 5′SS-25 (2:1 ratio) complexes revealed synthesis of near-full-length dystrophin compared with dystrophin extracted from normal C57BL muscle. No dystrophin protein was detected in muscle samples of untreated mdx mice.
Similar articles
- Specific removal of the nonsense mutation from the mdx dystrophin mRNA using antisense oligonucleotides.
Wilton SD, Lloyd F, Carville K, Fletcher S, Honeyman K, Agrawal S, Kole R. Wilton SD, et al. Neuromuscul Disord. 1999 Jul;9(5):330-8. doi: 10.1016/s0960-8966(99)00010-3. Neuromuscul Disord. 1999. PMID: 10407856 - Screening for antisense modulation of dystrophin pre-mRNA splicing.
Dickson G, Hill V, Graham IR. Dickson G, et al. Neuromuscul Disord. 2002 Oct;12 Suppl 1:S67-70. doi: 10.1016/s0960-8966(02)00085-8. Neuromuscul Disord. 2002. PMID: 12206799 Review. - Skipping multiple exons of dystrophin transcripts using cocktail antisense oligonucleotides.
Echigoya Y, Yokota T. Echigoya Y, et al. Nucleic Acid Ther. 2014 Feb;24(1):57-68. doi: 10.1089/nat.2013.0451. Epub 2013 Dec 31. Nucleic Acid Ther. 2014. PMID: 24380394 Review.
Cited by
- Splicing therapy for neuromuscular disease.
Douglas AG, Wood MJ. Douglas AG, et al. Mol Cell Neurosci. 2013 Sep;56:169-85. doi: 10.1016/j.mcn.2013.04.005. Epub 2013 Apr 28. Mol Cell Neurosci. 2013. PMID: 23631896 Free PMC article. Review. - Cell penetrating peptides: overview and applications to the delivery of oligonucleotides.
Said Hassane F, Saleh AF, Abes R, Gait MJ, Lebleu B. Said Hassane F, et al. Cell Mol Life Sci. 2010 Mar;67(5):715-26. doi: 10.1007/s00018-009-0186-0. Epub 2009 Nov 7. Cell Mol Life Sci. 2010. PMID: 19898741 Free PMC article. Review. - Repurposing Dantrolene for Long-Term Combination Therapy to Potentiate Antisense-Mediated DMD Exon Skipping in the mdx Mouse.
Wang DW, Mokhonova EI, Kendall GC, Becerra D, Naeini YB, Cantor RM, Spencer MJ, Nelson SF, Miceli MC. Wang DW, et al. Mol Ther Nucleic Acids. 2018 Jun 1;11:180-191. doi: 10.1016/j.omtn.2018.02.002. Epub 2018 Feb 13. Mol Ther Nucleic Acids. 2018. PMID: 29858053 Free PMC article. - Systematic evaluation of 2'-Fluoro modified chimeric antisense oligonucleotide-mediated exon skipping in vitro.
Chen S, Le BT, Chakravarthy M, Kosbar TR, Veedu RN. Chen S, et al. Sci Rep. 2019 Apr 15;9(1):6078. doi: 10.1038/s41598-019-42523-0. Sci Rep. 2019. PMID: 30988454 Free PMC article. - Improved cell-penetrating peptide-PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle.
Ivanova GD, Arzumanov A, Abes R, Yin H, Wood MJ, Lebleu B, Gait MJ. Ivanova GD, et al. Nucleic Acids Res. 2008 Nov;36(20):6418-28. doi: 10.1093/nar/gkn671. Epub 2008 Oct 8. Nucleic Acids Res. 2008. PMID: 18842625 Free PMC article.
References
- Koenig M, Monaco A P, Kunkel L M. Cell. 1988;53:219–228. - PubMed
- Hoffman E P, Brown R H, Jr, Kunkel L M. Cell. 1987;51:919–928. - PubMed
- Hodgson S V, Bobrow M. Br Med Bull. 1989;45:719–744. - PubMed
- Zatz M, Lange K, Spence M A. Lancet. 1977;1:759. - PubMed
- Monaco A P, Bertelson C J, Liechti-Gallati S, Moser H, Kunkel L M. Genomics. 1988;2:90–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases