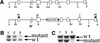Retention of PDGFR-beta function in mice in the absence of phosphatidylinositol 3'-kinase and phospholipase Cgamma signaling pathways - PubMed (original) (raw)
Retention of PDGFR-beta function in mice in the absence of phosphatidylinositol 3'-kinase and phospholipase Cgamma signaling pathways
M D Tallquist et al. Genes Dev. 2000.
Abstract
Signal transduction by the platelet-derived growth-factor receptor beta (PDGFR-beta) tyrosine kinase is required for proper formation of vascular smooth muscle cells (VSMC). However, the importance of individual PDGFR-beta signal transduction pathways in vivo is not known. To investigate the role of two of the pathways believed to be critical for PDGF signal transduction, we have generated mice that bear a PDGFR-beta that can no longer activate PI3kinase or PLCgamma. Although these mutant mice have normal vasculature, we provide multiple lines of evidence in vivo and from cells derived from the mutant mice that suggest that the mutant PDGFR-beta operates at suboptimal levels. Our observations indicate that although loss of these pathways can lead to attenuated PDGF-dependent cellular function, certain PDGFR-beta-induced signal cascades are not essential for survival in mice.
Figures
Figure 1
Generation of mice bearing a mutant PDGFR-β. (A) Wild type genomic and targeted locus. Region of targeting vector is flanked by shaded _Eco_RV (RV) and _Xho_I (X) sites. F indicates the exon where the Y ➞ F mutations (PI3kinase binding site) were made, and I indicates the exon where the Y ➞ I (PLCγ binding site) was made. (B) _Hin_dIII digest of targeted (lanes 1 and 2) and wild type (lane 3) ES cell clones. Probe P1 was used. Mutant band is at 6.9 kb, and wild type is at 5.2 kb. (C) _Eco_RV and _Asp_718 digest of same clones listed in B. Probe P2 was used. Mutant band is at 2.9 kb and wild type is at 4.8 kb. H, _Hin_dIII; RV, _Eco_RV; B, _Bsa_AI; S, _Spe_I; X, _Xho_I; A, _Asp_718. Triangles indicate loxP sites.
Figure 2
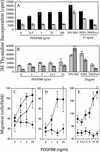
Expression and signaling of the mutant PDGFR-βF3/F3. Whole kidney lysates from mice (A) and cells (B) were blotted for protein levels. (A) Upper panel, PDGFR-β protein levels. Lower panel, RasGAP levels as a protein loading control. (B) Whole cell lysates from mesangial cells blotted for PDGFR-β protein levels. Lysates loaded in each lane are from 8 × 103 cells. (C–D) PLCγ phosphorylation. PLCγ was immunoprecipitated and blotted for tyrosine phosphorylation. The same filter was stripped and blotted for PLCγ. The lower band in the P-Tyr blot corresponds to PLCγ. (C) Wild type and F3M1 mesangial cells were stimulated with 30 ng/mL PDGF-BB. (D) Upper panels, mesangial cells were stimulated with the indicated concentration of PDGF-BB for 5 min and lysed. Lower panels, cells were stimulated with 30 ng/mL PDGF-BB for the indicated time and lysed. (E) IP3 assay. Cells were labeled o.n. with myo-[3H]inositol and then stimulated with PDGF-BB or 10% FBS. Cells were analyzed for inositol phosphate release. Data represent the mean and standard deviation of two independent experiments. Results are expressed as fold release, where the uninduced sample was used as the baseline and given the value of 1. In one experiment, 10% FBS was used as a positive control, where fold increase was as follows: WM1, 1.6; F3M1, 1.4; F3M2, 3.0; and F3/F3 MEFS, 1.4.
Figure 3
Lack of mitogenesis and migration in PDGFR-βF3/F3 cells. (A,B). [3H]thymidine incorporation of growth-factor-stimulated mouse mesangial cells. WM1 (A, solid black bars); F3M1 (A, B hatched bars); F3M2 (B, grey bars). Zero growth factor control was treated with vehicle alone. Each bar represents mean ± standard deviation of triplicate samples. Data representative of multiple independent assays. (C–E). Migration of MEFs and mesangial cells in a Neuroprobe chamber. Growth-factor-containing buffer was placed in the bottom well of the chamber. 5 × 104 MEFS or mesangial cells were placed in the top well and the cells were incubated for 5 hr. (C). Solid squares: PDGFR-α−/−/PDGFR-βF3/+ MEFs; solid circles: PDGFR-α−/−/PDGFR-βF3/F3 MEFs; solid triangles: PDGFR-α+/+/PDGFR-βF3/F3 MEFs. (D–E) Solid squares: Wild type mesangial cells (WM1); solid circles: F3 mesangial cells (F3M1 and F3M2, respectively). The number of migrated cells/well was counted in a field of C and D, 40× magnification or E, 20× magnification. Each data point represents the mean ± standard deviation of triplicate samples counted in 3–4 fields. Data are representative of multiple independent assays, except for MEFs, which were only assayed once.
Figure 4
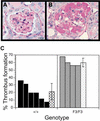
Quantitation of glomeruli-containing thrombi. (A,B) PAS-stained sections of anti-GBM-treated kidneys. (A) Glomerulus from a wild type diseased kidney with no thrombus. (B) Glomerulus from an F3 diseased kidney with thrombus formation. (C) Glomeruli were counted blindly for the presence or absence of a thrombus (as shown in A and B). Glomeruli 50–75 micron in diameter were included in the quantitation. Hatched bars indicate the average percentage of glomeruli that contained a thrombus ± standard deviation. No thrombi were observed in wild type or mutant mice that had been treated with normal sheep serum (NSS).
Figure 5
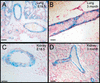
Distribution of cells in PDGFR-βF3/F3 ➞ R26+ wild type chimeras and PDGFR-β−/− ➞ R26+ wild type chimeras. (A) Coronal vibratome sections of hearts. Left, R26+ wild type heart (100%); middle, PDGFR-β−/− heart (20% wt chimera); right, PDGFR-βF3/F3 heart (28% wt chimera). (B) Vibratome sectioned tongue. Left, PDGFR-β−/− ➞ R26+ wild type (20% wt chimera); right, PDGFR-βF3/F3 ➞ R26+ wild type (28% wt chimera). (C) Hind limb vasculature from PDGFR-βF3/F3 chimerica (15% wt chimera). (D) Close-up of pulmonary artery from PDGFR-βF3/F3 chimeric lung (28% wt chimera). (E) Choroid plexus from PDGFR-βF3/F3 chimera (29% wt chimera). Arrows in all panels indicate concentrations of wild type cells in vessels. Percent chimerism was determined by Southern blot analysis.
Figure 6
Selection against F3-bearing cells in adults and neonates. (A,B) Lung section showing R26+ wild type cells in the vascular smooth muscle cell layers of the pulmonary arteries. (A) E18 chimeric embryo. (B) Adult chimera. (C,D) Arteries and arterioles in kidney of chimeric animals. (C) Arteriole from E18 chimeric embryo. (D) Arcuate artery from adult chimera. Note that the surrounding interstitial and tubule cells are mostly derived from F3 ES cells (not β-galactosidase positive) Bar: A,B, 50 microns; C,D, 25 microns.
Figure 7
Mesangial cells in the chimeric animals are wild type. (A) Sagittal vibratome section through a R26+ and chimeric E18.5 kidneys. Left, R26+ wild type kidney; middle, PDGFR-β−/− kidney; right, PDGFR-βF3/F3 kidney. 300 micron thickness. (B,C) Presence of β-galactosidase positive cells in the glomerulus of E18.5 kidneys while the surrounding tissue contains randomly scattered β-galactosidase positive cells. (D) Close-up of glomerulus and afferent artery in an adult chimera. (E) Quantitation of glomeruli that contain wild type cells. Black bars represent the percentage of wild type cells in each chimera as determined by quantitation of genomic tail DNA by Southern blot. Grey bars are the average percentage ± standard deviation of glomeruli that contain β-galactosidase positive cells. Four nonconsecutive sections from each chimera were used in the calculation. * Indicates that kidneys were from adult mice; all other analyses were from E18.5 embryos. F3-40 is a chimera that was generated from the ES cell line that had the neo gene deleted. Hatched boxes, chimeras generated using PDGFR-β null cells. Data are organized in descending percent of wild type chimerism as determined from tail DNA.
Similar articles
- Beta-platelet-derived growth factor receptor mediates motility and growth of Ewing's sarcoma cells.
Uren A, Merchant MS, Sun CJ, Vitolo MI, Sun Y, Tsokos M, Illei PB, Ladanyi M, Passaniti A, Mackall C, Toretsky JA. Uren A, et al. Oncogene. 2003 Apr 17;22(15):2334-42. doi: 10.1038/sj.onc.1206330. Oncogene. 2003. PMID: 12700668 - Platelet-derived growth factor-dependent cellular transformation requires either phospholipase Cgamma or phosphatidylinositol 3 kinase.
DeMali KA, Whiteford CC, Ulug ET, Kazlauskas A. DeMali KA, et al. J Biol Chem. 1997 Apr 4;272(14):9011-8. doi: 10.1074/jbc.272.14.9011. J Biol Chem. 1997. PMID: 9083025 - Platelet-derived growth factor stimulates protein kinase D through the activation of phospholipase Cgamma and protein kinase C.
Van Lint J, Ni Y, Valius M, Merlevede W, Vandenheede JR. Van Lint J, et al. J Biol Chem. 1998 Mar 20;273(12):7038-43. doi: 10.1074/jbc.273.12.7038. J Biol Chem. 1998. PMID: 9507012 - Nuclear phospholipase C and signaling.
Cocco L, Martelli AM, Gilmour RS, Rhee SG, Manzoli FA. Cocco L, et al. Biochim Biophys Acta. 2001 Jan 15;1530(1):1-14. doi: 10.1016/s1388-1981(00)00169-4. Biochim Biophys Acta. 2001. PMID: 11341954 Review. No abstract available. - The biology of platelet-derived growth factor.
Ross R, Raines EW, Bowen-Pope DF. Ross R, et al. Cell. 1986 Jul 18;46(2):155-69. doi: 10.1016/0092-8674(86)90733-6. Cell. 1986. PMID: 3013421 Review. No abstract available.
Cited by
- Novel mutations of PDGFRB cause primary familial brain calcification in Chinese families.
Wang C, Yao XP, Chen HT, Lai JH, Guo XX, Su HZ, Dong EL, Zhang QJ, Wang N, Chen WJ. Wang C, et al. J Hum Genet. 2017 Jul;62(7):697-701. doi: 10.1038/jhg.2017.25. Epub 2017 Mar 16. J Hum Genet. 2017. PMID: 28298627 - Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms.
Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Hamilton TG, et al. Mol Cell Biol. 2003 Jun;23(11):4013-25. doi: 10.1128/MCB.23.11.4013-4025.2003. Mol Cell Biol. 2003. PMID: 12748302 Free PMC article. - Functions and mechanisms of receptor tyrosine kinase Torso signaling: lessons from Drosophila embryonic terminal development.
Li WX. Li WX. Dev Dyn. 2005 Mar;232(3):656-72. doi: 10.1002/dvdy.20295. Dev Dyn. 2005. PMID: 15704136 Free PMC article. Review. - Vascular endothelial growth factors C and D induces proliferation of lymphangioleiomyomatosis cells through autocrine crosstalk with endothelium.
Issaka RB, Oommen S, Gupta SK, Liu G, Myers JL, Ryu JH, Vlahakis NE. Issaka RB, et al. Am J Pathol. 2009 Oct;175(4):1410-20. doi: 10.2353/ajpath.2009.080830. Epub 2009 Aug 28. Am J Pathol. 2009. PMID: 19717640 Free PMC article. - LRP1 regulates architecture of the vascular wall by controlling PDGFRbeta-dependent phosphatidylinositol 3-kinase activation.
Zhou L, Takayama Y, Boucher P, Tallquist MD, Herz J. Zhou L, et al. PLoS One. 2009 Sep 9;4(9):e6922. doi: 10.1371/journal.pone.0006922. PLoS One. 2009. PMID: 19742316 Free PMC article.
References
- Alimandi M, Heidaran MA, Gutkind JS, Zhang J, Ellmore N, Valius M, Kazlauskas A, Pierce JH, Li W. PLC-gamma activation is required for PDGF-βR-mediated mitogenesis and monocytic differentiation of myeloid progenitor cells. Oncogene. 1997;15:585–593. - PubMed
- Ataliotis P, Mercola M. Distribution and functions of platelet-derived growth factors and their receptors during embryogenesis. Int Rev Cytol. 1997;172:95–127. - PubMed
- Brückner K, Pasquale EB, Klein R. Tyrosine phosphorylation of transmembrane ligands for Eph receptors. Science. 1997;275:1640–1643. - PubMed
- Choudhury GG, Karamitsos C, Hernandez J, Gentilini A, Bardgette J, Abboud HE. PI-3-kinase and MAPK regulate mesangial cell proliferation and migration in response to PDGF. Amer J Phys. 1997;273:F931–938. - PubMed
- Crosby JR, Seifert RA, Soriano P, Bowen-Pope DF. Chimaeric analysis reveals role of Pdgf receptors in all muscle lineages. Nat Genet. 1998;18:385–388. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HD25326/HD/NICHD NIH HHS/United States
- R01 HD025326/HD/NICHD NIH HHS/United States
- R37 HD025326/HD/NICHD NIH HHS/United States
- HD24875/HD/NICHD NIH HHS/United States
- R01 HD024875/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous