The COG database: new developments in phylogenetic classification of proteins from complete genomes - PubMed (original) (raw)
The COG database: new developments in phylogenetic classification of proteins from complete genomes
R L Tatusov et al. Nucleic Acids Res. 2001.
Abstract
The database of Clusters of Orthologous Groups of proteins (COGs), which represents an attempt on a phylogenetic classification of the proteins encoded in complete genomes, currently consists of 2791 COGs including 45 350 proteins from 30 genomes of bacteria, archaea and the yeast Saccharomyces cerevisiae (http://www.ncbi.nlm.nih. gov/COG). In addition, a supplement to the COGs is available, in which proteins encoded in the genomes of two multicellular eukaryotes, the nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster, and shared with bacteria and/or archaea were included. The new features added to the COG database include information pages with structural and functional details on each COG and literature references, improvements of the COGNITOR program that is used to fit new proteins into the COGs, and classification of genomes and COGs constructed by using principal component analysis.
Figures
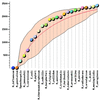
Figure 1
Growth dynamics of the COG set with the increase of number of included genomes. The circles show the sequence of genome inclusion according to the actual order of sequencing, and the smooth line shows the mean of 106 random permutations of the genome order. The colored area indicates the range between the maximal and minimal value for each point (number of genomes) in 106 random permutations.
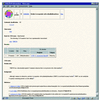
Figure 2
An example of a COG-Info page.
Figure 3
Classification of genome by co-occurrence in COGs using PCA. (A) All COGs. (B) Translation, transcription and replication (functional categories J, K and L). (C) Metabolism (functional categories C, E, F, G, H and I).
Similar articles
- The COG database: a tool for genome-scale analysis of protein functions and evolution.
Tatusov RL, Galperin MY, Natale DA, Koonin EV. Tatusov RL, et al. Nucleic Acids Res. 2000 Jan 1;28(1):33-6. doi: 10.1093/nar/28.1.33. Nucleic Acids Res. 2000. PMID: 10592175 Free PMC article. - The COG database: an updated version includes eukaryotes.
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. Tatusov RL, et al. BMC Bioinformatics. 2003 Sep 11;4:41. doi: 10.1186/1471-2105-4-41. Epub 2003 Sep 11. BMC Bioinformatics. 2003. PMID: 12969510 Free PMC article. - COG database update: focus on microbial diversity, model organisms, and widespread pathogens.
Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, Koonin EV. Galperin MY, et al. Nucleic Acids Res. 2021 Jan 8;49(D1):D274-D281. doi: 10.1093/nar/gkaa1018. Nucleic Acids Res. 2021. PMID: 33167031 Free PMC article. - Functional genomics and enzyme evolution. Homologous and analogous enzymes encoded in microbial genomes.
Galperin MY, Koonin EV. Galperin MY, et al. Genetica. 1999;106(1-2):159-70. doi: 10.1023/a:1003705601428. Genetica. 1999. PMID: 10710722 Review. - A genomic perspective on protein families.
Tatusov RL, Koonin EV, Lipman DJ. Tatusov RL, et al. Science. 1997 Oct 24;278(5338):631-7. doi: 10.1126/science.278.5338.631. Science. 1997. PMID: 9381173 Review.
Cited by
- Ubiquitous genome streamlined Acidobacteriota in freshwater environments.
Wong HL, Bulzu PA, Ghai R, Chiriac MC, Salcher MM. Wong HL, et al. ISME Commun. 2024 Oct 22;4(1):ycae124. doi: 10.1093/ismeco/ycae124. eCollection 2024 Jan. ISME Commun. 2024. PMID: 39544963 Free PMC article. - The coral Oculina patagonica holobiont and its response to confinement, temperature, and Vibrio infections.
Martin-Cuadrado AB, Rubio-Portillo E, Rosselló F, Antón J. Martin-Cuadrado AB, et al. Microbiome. 2024 Oct 29;12(1):222. doi: 10.1186/s40168-024-01921-x. Microbiome. 2024. PMID: 39472959 Free PMC article. - Complete genome sequence analysis and Pks genes identification of Brevibacillus brevis FJAT-0809-GLX with a broad inhibitory spectrum against phytopathogens.
Che J, Lai C, Lai G, Chen B, He L, Liu B. Che J, et al. World J Microbiol Biotechnol. 2024 Oct 3;40(11):332. doi: 10.1007/s11274-024-04139-z. World J Microbiol Biotechnol. 2024. PMID: 39358614 - Genomic Analysis of Antimicrobial Resistance in Pseudomonas aeruginosa from a "One Health" Perspective.
García-Rivera C, Molina-Pardines C, Haro-Moreno JM, Parra Grande M, Rodríguez JC, López-Pérez M. García-Rivera C, et al. Microorganisms. 2024 Aug 27;12(9):1770. doi: 10.3390/microorganisms12091770. Microorganisms. 2024. PMID: 39338445 Free PMC article. - Genomic and physiological properties of Anoxybacterium hadale gen. nov. sp. nov. reveal the important role of dissolved organic sulfur in microbial metabolism in hadal ecosystems.
Cao J, Shao B, Lin J, Liu J, Cui Y, Wang J, Fang J. Cao J, et al. Front Microbiol. 2024 Aug 16;15:1423245. doi: 10.3389/fmicb.2024.1423245. eCollection 2024. Front Microbiol. 2024. PMID: 39220043 Free PMC article.
References
- Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. - PubMed
- Fitch W.M. (1970) Distinguishing homologous from analogous proteins. Syst. Zool., 19, 99–106. - PubMed
- Kawarabayasi Y., Hino,Y., Horikawa,H., Yamazaki,S., Haikawa,Y., Jin-no,K., Takahashi,M., Sekine,M., Baba,S., Ankai,A. et al. (1999) Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res., 6, 83–101. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases