TIGRFAMs: a protein family resource for the functional identification of proteins - PubMed (original) (raw)
TIGRFAMs: a protein family resource for the functional identification of proteins
D H Haft et al. Nucleic Acids Res. 2001.
Abstract
TIGRFAMs is a collection of protein families featuring curated multiple sequence alignments, hidden Markov models and associated information designed to support the automated functional identification of proteins by sequence homology. We introduce the term 'equivalog' to describe members of a set of homologous proteins that are conserved with respect to function since their last common ancestor. Related proteins are grouped into equivalog families where possible, and otherwise into protein families with other hierarchically defined homology types. TIGRFAMs currently contains over 800 protein families, available for searching or downloading at www.tigr.org/TIGRFAMs. Classification by equivalog family, where achievable, complements classification by orthology, superfamily, domain or motif. It provides the information best suited for automatic assignment of specific functions to proteins from large-scale genome sequencing projects.
Figures
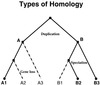
Figure 1
Homology relationships can be classified by evolutionary history, as shown in this model phylogenetic tree. The ancestral node, or root, is at the top. Duplication creates paralogs A and B with distinct function. Speciation creates an orthologous set A1, A2 and A3 from A, and B1, B2 and B3 from B. If B1, B2 and B3 share the same function, they are equivalogs as well as orthologs. Dashed lines indicate a possible pattern of gene loss that leaves only A1, B2 and B3. The resulting protein subfamily should exhibit bi-directional best hits across species but is not orthologous and does not show conserved function.
Similar articles
- The TIGRFAMs database of protein families.
Haft DH, Selengut JD, White O. Haft DH, et al. Nucleic Acids Res. 2003 Jan 1;31(1):371-3. doi: 10.1093/nar/gkg128. Nucleic Acids Res. 2003. PMID: 12520025 Free PMC article. - TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes.
Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. Selengut JD, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D260-4. doi: 10.1093/nar/gkl1043. Epub 2006 Dec 6. Nucleic Acids Res. 2007. PMID: 17151080 Free PMC article. - TIGRFAMs and Genome Properties in 2013.
Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. Haft DH, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D387-95. doi: 10.1093/nar/gks1234. Epub 2012 Nov 28. Nucleic Acids Res. 2013. PMID: 23197656 Free PMC article. - Database analysis and gene discovery in pharmacogenetics.
Board P, Tetlow N, Blackburn A, Chelvanayagam G. Board P, et al. Clin Chem Lab Med. 2000 Sep;38(9):863-7. doi: 10.1515/CCLM.2000.125. Clin Chem Lab Med. 2000. PMID: 11097341 Review. - Clustering and analysis of protein families.
Kriventseva EV, Biswas M, Apweiler R. Kriventseva EV, et al. Curr Opin Struct Biol. 2001 Jun;11(3):334-9. doi: 10.1016/s0959-440x(00)00211-6. Curr Opin Struct Biol. 2001. PMID: 11406384 Review.
Cited by
- De novo prediction of the genomic components and capabilities for microbial plant biomass degradation from (meta-)genomes.
Weimann A, Trukhina Y, Pope PB, Konietzny SG, McHardy AC. Weimann A, et al. Biotechnol Biofuels. 2013 Feb 15;6(1):24. doi: 10.1186/1754-6834-6-24. Biotechnol Biofuels. 2013. PMID: 23414703 Free PMC article. - Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence.
Thieme F, Koebnik R, Bekel T, Berger C, Boch J, Büttner D, Caldana C, Gaigalat L, Goesmann A, Kay S, Kirchner O, Lanz C, Linke B, McHardy AC, Meyer F, Mittenhuber G, Nies DH, Niesbach-Klösgen U, Patschkowski T, Rückert C, Rupp O, Schneiker S, Schuster SC, Vorhölter FJ, Weber E, Pühler A, Bonas U, Bartels D, Kaiser O. Thieme F, et al. J Bacteriol. 2005 Nov;187(21):7254-66. doi: 10.1128/JB.187.21.7254-7266.2005. J Bacteriol. 2005. PMID: 16237009 Free PMC article. - Finding New Cell Wall Regulatory Genes in Populus trichocarpa Using Multiple Lines of Evidence.
Furches A, Kainer D, Weighill D, Large A, Jones P, Walker AM, Romero J, Gazolla JGFM, Joubert W, Shah M, Streich J, Ranjan P, Schmutz J, Sreedasyam A, Macaya-Sanz D, Zhao N, Martin MZ, Rao X, Dixon RA, DiFazio S, Tschaplinski TJ, Chen JG, Tuskan GA, Jacobson D. Furches A, et al. Front Plant Sci. 2019 Oct 8;10:1249. doi: 10.3389/fpls.2019.01249. eCollection 2019. Front Plant Sci. 2019. PMID: 31649710 Free PMC article. - Classification of the plant-associated lifestyle of Pseudomonas strains using genome properties and machine learning.
Poncheewin W, van Diepeningen AD, van der Lee TAJ, Suarez-Diez M, Schaap PJ. Poncheewin W, et al. Sci Rep. 2022 Jun 27;12(1):10857. doi: 10.1038/s41598-022-14913-4. Sci Rep. 2022. PMID: 35760985 Free PMC article. - Complete Genome Sequence of the Extreme Thermophile Dictyoglomus thermophilum H-6-12.
Coil DA, Badger JH, Forberger HC, Riggs F, Madupu R, Fedorova N, Ward N, Robb FT, Eisen JA. Coil DA, et al. Genome Announc. 2014 Feb 20;2(1):e00109-14. doi: 10.1128/genomeA.00109-14. Genome Announc. 2014. PMID: 24558247 Free PMC article.
References
- Sonnhammer E.L., Eddy,S.R. and Durbin,R. (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins, 28, 405–420. - PubMed
- Srinivasarao G.Y., Yeh,L.S., Marzec,C.R., Orcutt,B.C. and Barker,W.C. (1999) PIR-ALN: a database of protein sequence alignments. Bioinformatics, 15, 382–390. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources