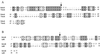The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV - PubMed (original) (raw)
The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV
F Qi et al. Appl Environ Microbiol. 2001 Jan.
Abstract
Strains of Streptococcus mutans produce at least three mutacins, I, II, and III. Mutacin II is a member of subgroup AII in the lantibiotic family of bacteriocins, and mutacins I and III belong to subgroup AI in the lantibiotic family. In this report, we characterize two mutacins produced by UA140, a group I strain of S. mutans. One is identical to the lantibiotic mutacin I produced by strain CH43 (F. Qi et al., Appl. Environ. Microbiol. 66:3221-3229, 2000); the other is a nonlantibiotic bacteriocin, which we named mutacin IV. Mutacin IV belongs to the two-peptide, nonlantibiotic family of bacteriocins produced by gram-positive bacteria. Peptide A, encoded by gene nlmA, is 44 amino acids (aa) in size and has a molecular mass of 4,169 Da; peptide B, encoded by nlmB, is 49 aa in size and has a molecular mass of 4,826 Da. Both peptides derive from prepeptides with glycines at positions -2 and -1 relative to the processing site. Production of mutacins I and IV by UA140 appears to be regulated by different mechanisms under different physiological conditions. The significance of producing two mutacins by one strain under different conditions and the implication of this property in terms of the ecology of S. mutans in the oral cavity are discussed.
Figures
FIG. 1
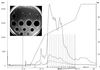
First-pass HPLC profile of crude mutacin extract from strain UA140 grown on a membrane. Fractions of 1 ml (dotted vertical lines) were collected during elution using buffers A (0.1% TFA) and B (0.085% TFA in 60% acetonitrile) and then tested for antimicrobial activity using NY101 as the indicator. The two active peaks, 1 (fractions 4 and 5) and 2 (fractions 8 and 9), are labeled. mAU, milli-absorption units.
FIG. 2
Product profile of UA140 grown in liquid culture under aerobic conditions. Crude mutacin extract was isolated and analyzed using the HPLC buffers and programs used for Fig. 1. Unlike the HPLC profile in Fig. 1, which showed two active peaks, only the second peak (fractions 7 to 14) was detected; the first peak (fraction 3) was barely detectable. mAU, milli-absorption units.
FIG. 3
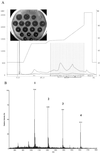
Purification and EIMS analysis of mutacin IV. (A) HPLC profile of the second pass for mutacin IV purification, using a different gradient to obtain better separation of different components. The active fractions were divided into three segments (fractions 7 to 13, 14 to 20, and 21 to 25) for EIMS analysis. (B) EIMS analysis of mutacin IV from fractions 7 to 13 in panel A. Peaks 1 and 3 correspond to quadruply and triply charged molecules of 4,169 Da, respectively; peaks 2 and 4 correspond to quadruply and triply charged molecules of 4,826 Da, respectively. mAU, milli-absorption units.
FIG. 4
(A) Genomic organization of the mutacin IV gene locus. The genes encoding proteins with similarity to known enzymes or proteins are labeled. nlmA and nlmB are structural genes for the mutacin IV prepeptides. (B) DNA and deduced amino acid sequences of the two ORFs in contig 450. Bold letters in Orf1 and Orf2 correspond to peptide sequences obtained by N-terminal sequencing of the two peptides in mutacin IV. Boxed letters are the double glycines at the prepeptide cleavage site. The putative ribosomal binding sites for orf1 and orf2 are underlined.
FIG. 5
Similarity of NlmA and NlmB with other peptides in GenBank. (A) Sequence alignment of NlmA and ThmA; (B) sequence alignment of NlmB and LafA. ThmA and LafA are each one of two components in the two-peptide nonlantibiotic bacteriocin thermophilin 13 and lactacin F, respectively. Dark gray boxes represent identical amino acids; light gray boxes denote conserved changes; arrows indicate cleavage sites for the prepeptide.
Similar articles
- Production, purification, sequencing and activity spectra of mutacins D-123.1 and F-59.1.
Nicolas GG, LaPointe G, Lavoie MC. Nicolas GG, et al. BMC Microbiol. 2011 Apr 10;11:69. doi: 10.1186/1471-2180-11-69. BMC Microbiol. 2011. PMID: 21477375 Free PMC article. - Genetic Analysis of Mutacin B-Ny266, a Lantibiotic Active against Caries Pathogens.
Dufour D, Barbour A, Chan Y, Cheng M, Rahman T, Thorburn M, Stewart C, Finer Y, Gong SG, Lévesque CM. Dufour D, et al. J Bacteriol. 2020 May 27;202(12):e00762-19. doi: 10.1128/JB.00762-19. Print 2020 May 27. J Bacteriol. 2020. PMID: 32229530 Free PMC article. - The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc.
Chen P, Qi F, Novak J, Caufield PW. Chen P, et al. Appl Environ Microbiol. 1999 Mar;65(3):1356-60. doi: 10.1128/AEM.65.3.1356-1360.1999. Appl Environ Microbiol. 1999. PMID: 10049909 Free PMC article. - The mutacins of Streptococcus mutans: regulation and ecology.
Merritt J, Qi F. Merritt J, et al. Mol Oral Microbiol. 2012 Apr;27(2):57-69. doi: 10.1111/j.2041-1014.2011.00634.x. Epub 2011 Dec 23. Mol Oral Microbiol. 2012. PMID: 22394465 Free PMC article. Review. - [Bacteriocins of Streptococcus mutans: mutacins].
Baca Garcia P, Liebana Ureña J. Baca Garcia P, et al. Rev Eur Odontoestomatol. 1990 Mar-Apr;2(2):127-30. Rev Eur Odontoestomatol. 1990. PMID: 2222663 Review. Spanish.
Cited by
- investigating acid production by Streptococcus mutans with a surface-displayed pH-sensitive green fluorescent protein.
Guo L, Hu W, He X, Lux R, McLean J, Shi W. Guo L, et al. PLoS One. 2013;8(2):e57182. doi: 10.1371/journal.pone.0057182. Epub 2013 Feb 28. PLoS One. 2013. PMID: 23468929 Free PMC article. - Recent Advances in Screening of Anti-Campylobacter Activity in Probiotics for Use in Poultry.
Saint-Cyr MJ, Guyard-Nicodème M, Messaoudi S, Chemaly M, Cappelier JM, Dousset X, Haddad N. Saint-Cyr MJ, et al. Front Microbiol. 2016 May 31;7:553. doi: 10.3389/fmicb.2016.00553. eCollection 2016. Front Microbiol. 2016. PMID: 27303366 Free PMC article. Review. - Interspecies interactions within oral microbial communities.
Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Kuramitsu HK, et al. Microbiol Mol Biol Rev. 2007 Dec;71(4):653-70. doi: 10.1128/MMBR.00024-07. Microbiol Mol Biol Rev. 2007. PMID: 18063722 Free PMC article. Review. - Regulation of ciaXRH operon expression and identification of the CiaR regulon in Streptococcus mutans.
Wu C, Ayala EA, Downey JS, Merritt J, Goodman SD, Qi F. Wu C, et al. J Bacteriol. 2010 Sep;192(18):4669-79. doi: 10.1128/JB.00556-10. Epub 2010 Jul 16. J Bacteriol. 2010. PMID: 20639331 Free PMC article. - Complete Genome Sequences of Two Mutacin-Producing Streptococcus mutans Strains, T8 and UA140.
Biswas I. Biswas I. Microbiol Resour Announc. 2020 Jun 11;9(24):e00469-20. doi: 10.1128/MRA.00469-20. Microbiol Resour Announc. 2020. PMID: 32527777 Free PMC article.
References
- Balakrishnan M, Simmonds R S, Carne A, Tagg J R. Streptococcus mutans strain N produces a novel low molecular mass non-lantibiotic bacteriocin. FEMS Microbiol Lett. 2000;183:165–169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical