Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in reduction of manganese dioxide - PubMed (original) (raw)
Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in reduction of manganese dioxide
J M Myers et al. Appl Environ Microbiol. 2001 Jan.
Abstract
Shewanella putrefaciens MR-1 can use a wide variety of terminal electron acceptors for anaerobic respiration, including certain insoluble manganese and iron oxides. To examine whether the outer membrane (OM) cytochromes of MR-1 play a role in Mn(IV) and Fe(III) reduction, mutants lacking the OM cytochrome OmcA or OmcB were isolated by gene replacement. Southern blotting and PCR confirmed replacement of the omcA and omcB genes, respectively, and reverse transcription-PCR analysis demonstrated loss of the respective mRNAs, whereas mRNAs for upstream and downstream genes were retained. The omcA mutant (OMCA1) resembled MR-1 in its growth on trimethylamine N-oxide (TMAO), dimethyl sulfoxide, nitrate, fumarate, thiosulfate, and tetrathionate and its reduction of nitrate, nitrite, ferric citrate, FeOOH, and anthraquinone-2,6-disulfonic acid. Similarly, the omcB mutant (OMCB1) grew on fumarate, nitrate, TMAO, and thiosulfate and reduced ferric citrate and FeOOH. However, OMCA1 and OMCB1 were 45 and 75% slower than MR-1, respectively, at reducing MnO(2). OMCA1 lacked only OmcA. While OMCB1 lacked OmcB, other OM cytochromes were also missing or markedly depressed. The total cytochrome content of the OM of OMCB1 was less than 15% of that of MR-1. Western blots demonstrated that OMCB1 still synthesized OmcA, but most of it was localized in the cytoplasmic membrane and soluble fractions rather than in the OM. OMCB1 had therefore lost the ability to properly localize multiple OM cytochromes to the OM. Together, the results suggest that the OM cytochromes of MR-1 participate in the reduction of Mn(IV) but are not required for the reduction of Fe(III) or other electron acceptors.
Figures
FIG. 1
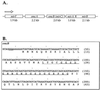
(A) Linear orientation of the gene cluster of MR-1 surrounding omcA and omcB as derived from GenBank accession no. AF083240 (2). The diagram is not drawn to scale. The transcription direction in all cases is from left to right (5′ → 3′). (B) N-terminal sequencing of purified OmcB yielded the underlined amino acid sequence, which corresponded exactly to the predicted coding region of omcB (only 200 bp of DNA sequence are shown, beginning just upstream of the omcB start codon). The cysteine at residue 25 is likely the first residue of mature OmcB but was unidentified during N-terminal sequencing, consistent with the putative lipoprotein modification of this residue. The lipoprotein consensus sequence (LTGC) is indicated by the double underline. The numbers at right indicate arbitrary positions within the nucleotide sequence (top) and absolute numbers of amino acid residues (bottom), with 1 corresponding to the N terminus of the immature protein.
FIG. 2
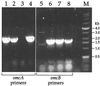
PCR products with genomic DNA templates from the following strains: lanes 1, OMCB1; lanes 2, OMCA1; lanes 3, MR-1. The following primer pairs (Table 2) were used: B1 and B2 for omcB (A), A1 and A2 for omcA (B), and M1 and M2 for mtrA-mtrB (C).
FIG. 3
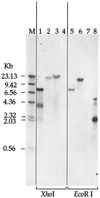
Southern blot of genomic DNA from Shewanella strains or of pUT/Tn_5Km_ plasmid DNA probed with the Kmr gene from pUT/Tn_5Km_. The lanes were loaded with DNA that was digested with either _Xho_I (lanes 1 to 4) or Eco_RI (lanes 5 to 8). Lanes 1 and 8, pUT/Tn_5Km; lanes 2 and 6, OMCA1; lanes 3 and 5, OMCB1; lanes 4 and 7, MR-1. The sizes of the DNA markers (lane M) are indicated on the left.
FIG. 4
RT-PCR products using total RNA from the following strains (grown anaerobically with fumarate) as templates: lanes 1 and 5, OMCB1; lanes 2, 4, 6, and 8, MR-1; lanes 3 and 7, OMCA1. Primers A5 and A6 (Table 2), specific for omcA, were used for the RT-PCRs in lanes 1 to 4, and primers B5 and B6, specific for omcB, were used for the reactions in lanes 5 to 8. The sizes of the DNA markers (lane M) are indicated on the right. Controls without reverse transcriptase yielded no bands (not shown), indicating that the products shown arise from mRNA.
FIG. 5
SDS-PAGE of subcellular fractions prepared from MR-1 (lanes 1, 3, 5, and 7) or OMCA1 (lanes 2, 4, 6, and 8) cells grown anaerobically with fumarate as the electron acceptor. (A) Gel stained for heme. (B) Western blot of a duplicate of the gel in panel A run under identical conditions and probed with an IgG specific for OmcA (24). The lanes were loaded with 10 (A) or 5 (B) μg of protein from each of the following subcellular fractions: CM (lanes 1 and 2), IM (lanes 3 and 4), OM (lanes 5 and 6), and soluble fraction (lanes 7 and 8). OMCA1 lacks the band corresponding to OmcA (open arrows), but it retains the OmcB protein (solid arrows). The Western blot demonstrates the presence of OmcA in MR-1 and its absence in OMCA1. The bars and numbers at the left indicate the migration and masses of the protein standards obtained from a parallel gel containing the same samples but stained for protein.
FIG. 6
Heme-stained SDS-PAGE profiles of subcellular fractions prepared from MR-1/pVK100 (lanes 1, 4, 7, and 10), OMCB2 (lanes 2, 5, 8, and 11), or OMCB1/pVK100 (lanes 3, 6, 9, and 12) cells grown anaerobically with fumarate as the electron acceptor. The lanes were loaded with 25 μg of protein from each of the following subcellular fractions: CM (lanes 1 to 3), IM (lanes 4 to 6), OM (lanes 7 to 9), and soluble fraction (lanes 10 to 12). Strains OMCB1 and OMCB2 lack the band corresponding to OmcB (arrows) in the IM and OM. The bars and numbers at the left indicate the migration and masses of the protein standards obtained from a parallel gel containing the same samples but stained for protein.
FIG. 7
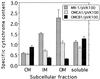
Specific cytochrome content of subcellular fractions prepared from the indicated strains, which were grown anaerobically with fumarate as the electron acceptor. The specific cytochrome content is the difference between the absorbances at the peak and trough of the Soret region from reduced-minus-oxidized difference spectra per milligram of protein. The values represent means plus the high values for two parallel but independent experiments.
FIG. 8
Western blot using polyclonal IgG specific for OmcA. The lanes were loaded with 5 μg of protein from each of the following subcellular fractions: CM (lanes 1 to 3), IM (lanes 4 to 6), OM (lanes 7 to 9), and soluble fraction (lanes 10 to 12). The subcellular fractions were prepared from fumarate-grown cells of the following strains: MR-1/pVK100 (lanes 1, 4, 7, and 10), OMCB1/pVK100 (lanes 2, 5, 8, and 11), and OMCA1/pVK100 (lanes 3, 6, 9, and 12).
FIG. 9
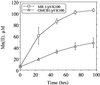
Reduction of δMnO2 by MR-1 and OMCB1 under anaerobic conditions as determined by the formation of Mn(II) over time. The points represent the means for an n of 2, and the bars represent the range of high and low values; for points lacking apparent range bars, the bars were smaller than the points as shown.
Similar articles
- Genetic complementation of an outer membrane cytochrome omcB mutant of Shewanella putrefaciens MR-1 requires omcB plus downstream DNA.
Myers JM, Myers CR. Myers JM, et al. Appl Environ Microbiol. 2002 Jun;68(6):2781-93. doi: 10.1128/AEM.68.6.2781-2793.2002. Appl Environ Microbiol. 2002. PMID: 12039733 Free PMC article. - MtrB is required for proper incorporation of the cytochromes OmcA and OmcB into the outer membrane of Shewanella putrefaciens MR-1.
Myers CR, Myers JM. Myers CR, et al. Appl Environ Microbiol. 2002 Nov;68(11):5585-94. doi: 10.1128/AEM.68.11.5585-5594.2002. Appl Environ Microbiol. 2002. PMID: 12406753 Free PMC article. - Overlapping role of the outer membrane cytochromes of Shewanella oneidensis MR-1 in the reduction of manganese(IV) oxide.
Myers JM, Myers CR. Myers JM, et al. Lett Appl Microbiol. 2003;37(1):21-5. doi: 10.1046/j.1472-765x.2003.01338.x. Lett Appl Microbiol. 2003. PMID: 12803550 - Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes.
Shi L, Squier TC, Zachara JM, Fredrickson JK. Shi L, et al. Mol Microbiol. 2007 Jul;65(1):12-20. doi: 10.1111/j.1365-2958.2007.05783.x. Mol Microbiol. 2007. PMID: 17581116 Free PMC article. Review. - Dissimilatory Fe(III) and Mn(IV) reduction.
Lovley DR, Holmes DE, Nevin KP. Lovley DR, et al. Adv Microb Physiol. 2004;49:219-86. doi: 10.1016/S0065-2911(04)49005-5. Adv Microb Physiol. 2004. PMID: 15518832 Review.
Cited by
- Iron-oxide minerals affect extracellular electron-transfer paths of Geobacter spp.
Kato S, Hashimoto K, Watanabe K. Kato S, et al. Microbes Environ. 2013;28(1):141-8. doi: 10.1264/jsme2.me12161. Epub 2013 Jan 30. Microbes Environ. 2013. PMID: 23363619 Free PMC article. - Isolation, characterization and gene sequence analysis of a membrane-associated 89 kDa Fe(III) reducing cytochrome c from Geobacter sulfurreducens.
Magnuson TS, Isoyama N, Hodges-Myerson AL, Davidson G, Maroney MJ, Geesey GG, Lovley DR. Magnuson TS, et al. Biochem J. 2001 Oct 1;359(Pt 1):147-52. doi: 10.1042/0264-6021:3590147. Biochem J. 2001. PMID: 11563978 Free PMC article. - Role of multiheme cytochromes involved in extracellular anaerobic respiration in bacteria.
Edwards MJ, Richardson DJ, Paquete CM, Clarke TA. Edwards MJ, et al. Protein Sci. 2020 Apr;29(4):830-842. doi: 10.1002/pro.3787. Epub 2019 Nov 28. Protein Sci. 2020. PMID: 31721352 Free PMC article. Review. - Purification and characterization of the [NiFe]-hydrogenase of Shewanella oneidensis MR-1.
Shi L, Belchik SM, Plymale AE, Heald S, Dohnalkova AC, Sybirna K, Bottin H, Squier TC, Zachara JM, Fredrickson JK. Shi L, et al. Appl Environ Microbiol. 2011 Aug 15;77(16):5584-90. doi: 10.1128/AEM.00260-11. Epub 2011 Jul 1. Appl Environ Microbiol. 2011. PMID: 21724888 Free PMC article. - Recent Developments and Applications of Microbial Electrochemical Biosensors.
Carducci NGG, Dey S, Hickey DP. Carducci NGG, et al. Adv Biochem Eng Biotechnol. 2024;187:149-183. doi: 10.1007/10_2023_236. Adv Biochem Eng Biotechnol. 2024. PMID: 38273205 Review.
References
- Armstrong P B, Lyons W B, Gaudette H E. Application of formaldoxime colorimetric method for the determination of manganese in the pore water of anoxic estuarine sediments. Estuaries. 1979;2:198–201.
- Braun V. Energy-coupled transport and signal transduction through the Gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. - PubMed
- Brewer P G, Spencer D W. Colorimetric determination of manganese in anoxic waters. Limnol Oceanogr. 1971;16:107–110.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases