Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives - PubMed (original) (raw)
Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives
P Hugenholtz et al. Appl Environ Microbiol. 2001 Jan.
Abstract
A molecular approach was used to investigate a recently described candidate division of the domain Bacteria, TM7, currently known only from environmental 16S ribosomal DNA sequence data. A number of TM7-specific primers and probes were designed and evaluated. Fluorescence in situ hybridization (FISH) of a laboratory scale bioreactor using two independent TM7-specific probes revealed a conspicuous sheathed-filament morphotype, fortuitously enriched in the reactor. Morphologically, the filament matched the description of the Eikelboom morphotype 0041-0675 widely associated with bulking problems in activated-sludge wastewater treatment systems. Transmission electron microscopy of the bioreactor sludge demonstrated that the sheathed-filament morphotype had a typical gram-positive cell envelope ultrastructure. Therefore, TM7 is only the third bacterial lineage recognized to have gram-positive representatives. TM7-specific FISH analysis of two full-scale wastewater treatment plant sludges, including the one used to seed the laboratory scale reactor, indicated the presence of a number of morphotypes, including sheathed filaments. TM7-specific PCR clone libraries prepared from the two full-scale sludges yielded 23 novel TM7 sequences. Three subdivisions could be defined based on these data and publicly available sequences. Environmental sequence data and TM7-specific FISH analysis indicate that members of the TM7 division are present in a variety of terrestrial, aquatic, and clinical habitats. A highly atypical base substitution (Escherichia coli position 912; C to U) for bacterial 16S rRNAs was present in almost all TM7 sequences, suggesting that TM7 bacteria, like Archaea, may be streptomycin resistant at the ribosome level.
Figures
FIG. 1
Evolutionary-distance dendrogram of candidate division TM7 and other bacterial division level groups based on comparative analyses of 16S rDNA data. Division and subdivision designations are bracketed on the right. Specificities of primers and probes designed to target the TM7 division (detailed in Table 1) are indicated in italics outside the brackets. Branch points supported (bootstrap values, >74%) by all inference methods used are indicated by solid circles, and those supported by most inference methods are indicated by open circles. Branch points without circles were not resolved (bootstrap values, <75%) as specific groups in different analyses and at the division level were collapsed back to the next significant node. Clones sequenced in the present study are in boldface. Partial-length TM7 sequences (<600 nt) were inserted into the tree using the parsimony insertion tool of ARB to show their approximate positions and are indicated by dashed line segments. Archaeal outgroups (not shown) for the tree were Methanococcus vannielii (M36507) and Sulfolobus acidocaldarius (D14876). The bar represents 10% estimated sequence divergence.
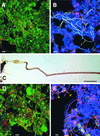
FIG. 2
Light microscopy of the microbial communities of laboratory scale and full-scale activated-sludge samples. All bars are 10 μm, except for that in panel C, which is 5 μm. (A and D) Confocal laser scanning microscopy (CLSM) images of a laboratory scale EBPR sludge (A) and Loganholme wastewater treatment plant (WTP) sludge (D) dual hybridized with FITC-labeled EUB338 and CY3-labeled TM7305. TM7 morphotypes appear yellow-orange, and other bacteria appear green. (C) Bright-field micrograph of a Gram stain of the sheathed-filament morphotype in the laboratory scale sludge. Note the sheath and prominent cuff. (B and E) CLSM images of the laboratory scale EBPR sludge (B) and Noosa WTP sludge (E) triple hybridized with FITC-labeled TM7305, CY3-labeled TM7905, and CY5-labeled EUB338. TM7 morphotypes which bind both TM7-specific probes and EUB338 appear white, morphotypes which bind only TM7905 and EUB338 appear pink, and other non-TM7 bacteria appear blue. Selected TM7 morphotypes are highlighted with arrows. f, filament; c, cocci; r, rod; tf, thick filament.
FIG. 3
Transmission electron micrographs of TM7 sheathed-filament morphotype. (A and B) Longitudinal sections of filament showing pronounced sheath (S), attached to the filament at one point (A), and septum (SE). Bars = 0.5 μm (A) and 100 nm (B). (C) High-magnification image of a cross section of a cell showing typical gram-positive cell envelope, cytoplasmic membrane (CM), and outer wall (W). Note the trilaminar appearance of the wall. Bar = 50 nm. (D) Cross section of a filament showing a series of fine ridges (R) running longitudinally along the cells, the sheath (S), and close association of the sheath with the filament (A). Bar = 100 nm. (E) Filament terminus showing a sheath cuff extending beyond the cells. Bar = 0.5 μm.
FIG. 4
Consensus sequences of part of 16S rRNA stem 30 (according to the ARB numbering system) in Bacteria (except TM7), Archaea, and TM7. The residue at position 912 (circled) is primarily responsible for streptomycin resistance (Smr) and sensitivity (Sms) in different domains (3). Canonical and noncanonical base pairing between residues are indicated by lines and dots, respectively.
Similar articles
- Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester.
Ariesyady HD, Ito T, Okabe S. Ariesyady HD, et al. Water Res. 2007 Apr;41(7):1554-68. doi: 10.1016/j.watres.2006.12.036. Epub 2007 Feb 8. Water Res. 2007. PMID: 17291558 - Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester.
Chouari R, Le Paslier D, Daegelen P, Ginestet P, Weissenbach J, Sghir A. Chouari R, et al. Environ Microbiol. 2005 Aug;7(8):1104-15. doi: 10.1111/j.1462-2920.2005.00795.x. Environ Microbiol. 2005. PMID: 16011748 - Fluorescence in situ hybridization probes targeting members of the phylum Candidatus Saccharibacteria falsely target Eikelboom type 1851 filaments and other Chloroflexi members.
Nittami T, Speirs LB, Fukuda J, Watanabe M, Seviour RJ. Nittami T, et al. Environ Microbiol Rep. 2014 Dec;6(6):611-7. doi: 10.1111/1758-2229.12172. Environ Microbiol Rep. 2014. PMID: 25756114 - A survey of the relative abundance of specific groups of cellulose degrading bacteria in anaerobic environments using fluorescence in situ hybridization.
O'Sullivan C, Burrell PC, Clarke WP, Blackall LL. O'Sullivan C, et al. J Appl Microbiol. 2007 Oct;103(4):1332-43. doi: 10.1111/j.1365-2672.2007.03362.x. J Appl Microbiol. 2007. PMID: 17897237 Review. - A phylum level perspective on bacterial cell envelope architecture.
Sutcliffe IC. Sutcliffe IC. Trends Microbiol. 2010 Oct;18(10):464-70. doi: 10.1016/j.tim.2010.06.005. Epub 2010 Jul 14. Trends Microbiol. 2010. PMID: 20637628 Review.
Cited by
- Globally distributed marine Gemmatimonadota have unique genomic potentials.
Gong X, Xu L, Langwig MV, Chen Z, Huang S, Zhao D, Su L, Zhang Y, Francis CA, Liu J, Li J, Baker BJ. Gong X, et al. Microbiome. 2024 Aug 10;12(1):149. doi: 10.1186/s40168-024-01871-4. Microbiome. 2024. PMID: 39123272 Free PMC article. - Episymbiotic Saccharibacteria induce intracellular lipid droplet production in their host bacteria.
Dong PT, Tian J, Kobayashi-Kirschvink KJ, Cen L, McLean JS, Bor B, Shi W, He X. Dong PT, et al. ISME J. 2024 Jan 8;18(1):wrad034. doi: 10.1093/ismejo/wrad034. ISME J. 2024. PMID: 38366018 Free PMC article. - The oral microbiome: diversity, biogeography and human health.
Baker JL, Mark Welch JL, Kauffman KM, McLean JS, He X. Baker JL, et al. Nat Rev Microbiol. 2024 Feb;22(2):89-104. doi: 10.1038/s41579-023-00963-6. Epub 2023 Sep 12. Nat Rev Microbiol. 2024. PMID: 37700024 Free PMC article. Review. - Metallo-Beta-Lactamase-like Encoding Genes in Candidate Phyla Radiation: Widespread and Highly Divergent Proteins with Potential Multifunctionality.
Maatouk M, Merhej V, Pontarotti P, Ibrahim A, Rolain JM, Bittar F. Maatouk M, et al. Microorganisms. 2023 Jul 28;11(8):1933. doi: 10.3390/microorganisms11081933. Microorganisms. 2023. PMID: 37630493 Free PMC article. - Distribution, abundance, and ecogenomics of the Palauibacterales, a new cosmopolitan thiamine-producing order within the Gemmatimonadota phylum.
Aldeguer-Riquelme B, Antón J, Santos F. Aldeguer-Riquelme B, et al. mSystems. 2023 Aug 31;8(4):e0021523. doi: 10.1128/msystems.00215-23. Epub 2023 Jun 22. mSystems. 2023. PMID: 37345931 Free PMC article.
References
- Amils R, Cammarano P, Londei P. Translation in archaea. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of archaea (archaebacteria) Vol. 26. Amsterdam, The Netherlands: Elsevier; 1993. pp. 393–438.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases