Multilocus sequence typing system for Campylobacter jejuni - PubMed (original) (raw)
Multilocus sequence typing system for Campylobacter jejuni
K E Dingle et al. J Clin Microbiol. 2001 Jan.
Abstract
The gram-negative bacterium Campylobacter jejuni has extensive reservoirs in livestock and the environment and is a frequent cause of gastroenteritis in humans. To date, the lack of (i) methods suitable for population genetic analysis and (ii) a universally accepted nomenclature has hindered studies of the epidemiology and population biology of this organism. Here, a multilocus sequence typing (MLST) system for this organism is described, which exploits the genetic variation present in seven housekeeping loci to determine the genetic relationships among isolates. The MLST system was established using 194 C. jejuni isolates of diverse origins, from humans, animals, and the environment. The allelic profiles, or sequence types (STs), of these isolates were deposited on the Internet (http://mlst.zoo.ox.ac.uk), forming a virtual isolate collection which could be continually expanded. These data indicated that C. jejuni is genetically diverse, with a weakly clonal population structure, and that intra- and interspecies horizontal genetic exchange was common. Of the 155 STs observed, 51 (26% of the isolate collection) were unique, with the remainder of the collection being categorized into 11 lineages or clonal complexes of related STs with between 2 and 56 members. In some cases membership in a given lineage or ST correlated with the possession of a particular Penner HS serotype. Application of this approach to further isolate collections will enable an integrated global picture of C. jejuni epidemiology to be established and will permit more detailed studies of the population genetics of this organism.
Figures
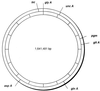
FIG. 1
Chromosomal locations of MLST loci. The positions of the seven loci are shown on a map of the C. jejuni chromosome derived from the genome sequence of isolate NCTC 11168 (
http://www.sanger.ac.uk/Projects/C\_jejuni/
). The 1,641,481-bp genome is divided into 10 segments (indicated on the inner circle), with each segment representing 164,148 bp.
FIG. 2
Allele frequencies in the sample population. For each of the seven loci (A to G), the number of times that each allele occurs in the isolate collection is shown. The frequencies are shown in the order of most to least abundant.
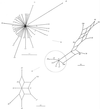
FIG. 3
Splits graphs showing the interrelationships of members of ST-21 complex. Progressive pruning of branches to improve resolution is indicated by the circles and arrows. The completely resolved region (bottom left) derived from the center of the unpruned graph contains the most abundant members of the complex and the predicted founder of the lineage, ST-21.
Similar articles
- Multilocus sequence typing of Campylobacter jejuni from broilers.
Griekspoor P, Engvall EO, Olsen B, Waldenström J. Griekspoor P, et al. Vet Microbiol. 2010 Jan 6;140(1-2):180-5. doi: 10.1016/j.vetmic.2009.07.022. Epub 2009 Aug 8. Vet Microbiol. 2010. PMID: 19733453 - Genetic diversity among Campylobacter jejuni isolates from healthy livestock and their links to human isolates in Spain.
Oporto B, Juste RA, López-Portolés JA, Hurtado A. Oporto B, et al. Zoonoses Public Health. 2011 Aug;58(5):365-75. doi: 10.1111/j.1863-2378.2010.01373.x. Epub 2010 Oct 7. Zoonoses Public Health. 2011. PMID: 21040505 - Allelic diversity and recombination in Campylobacter jejuni.
Suerbaum S, Lohrengel M, Sonnevend A, Ruberg F, Kist M. Suerbaum S, et al. J Bacteriol. 2001 Apr;183(8):2553-9. doi: 10.1128/JB.183.8.2553-2559.2001. J Bacteriol. 2001. PMID: 11274115 Free PMC article. - Campylobacter sequence typing databases: applications and future prospects.
Colles FM, Maiden MCJ. Colles FM, et al. Microbiology (Reading). 2012 Nov;158(Pt 11):2695-2709. doi: 10.1099/mic.0.062000-0. Epub 2012 Sep 17. Microbiology (Reading). 2012. PMID: 22986295 Review. - Genetic differentiation of Campylobacter jejuni.
Wassenaar TM. Wassenaar TM. Int J Infect Dis. 2002 Dec;6 Suppl 3:3S22-5; discussion 3S25, 3S53-8. doi: 10.1016/s1201-9712(02)90180-3. Int J Infect Dis. 2002. PMID: 23570170 Review.
Cited by
- Antimicrobial Resistance and Virulence-Associated Markers in Campylobacter Strains From Diarrheic and Non-diarrheic Humans in Poland.
Wysok B, Wojtacka J, Hänninen ML, Kivistö R. Wysok B, et al. Front Microbiol. 2020 Aug 4;11:1799. doi: 10.3389/fmicb.2020.01799. eCollection 2020. Front Microbiol. 2020. PMID: 32849410 Free PMC article. - Genomic Analysis Points to Multiple Genetic Mechanisms for Non-Transformable Campylobacter jejuni ST-50.
Parker CT, Villafuerte DA, Miller WG, Huynh S, Chapman MH, Hanafy Z, Jackson JH 3rd, Miller MA, Kathariou S. Parker CT, et al. Microorganisms. 2024 Feb 4;12(2):327. doi: 10.3390/microorganisms12020327. Microorganisms. 2024. PMID: 38399730 Free PMC article. - Partial Failure of Milk Pasteurization as a Risk for the Transmission of Campylobacter From Cattle to Humans.
Fernandes AM, Balasegaram S, Willis C, Wimalarathna HM, Maiden MC, McCarthy ND. Fernandes AM, et al. Clin Infect Dis. 2015 Sep 15;61(6):903-9. doi: 10.1093/cid/civ431. Epub 2015 Jun 10. Clin Infect Dis. 2015. PMID: 26063722 Free PMC article. - The prevalence of Campylobacter amongst a free-range broiler breeder flock was primarily affected by flock age.
Colles FM, McCarthy ND, Layton R, Maiden MC. Colles FM, et al. PLoS One. 2011;6(12):e22825. doi: 10.1371/journal.pone.0022825. Epub 2011 Dec 12. PLoS One. 2011. PMID: 22174732 Free PMC article. - Prevalence of thermophilic Campylobacter species in Swedish dogs and characterization of C. jejuni isolates.
Holmberg M, Rosendal T, Engvall EO, Ohlson A, Lindberg A. Holmberg M, et al. Acta Vet Scand. 2015 Apr 1;57(1):19. doi: 10.1186/s13028-015-0108-0. Acta Vet Scand. 2015. PMID: 25884591 Free PMC article.
References
- Bolton F J, Wareing D R A, Skirrow M B, Hutchinson D N. Identification and biotyping of campylobacters. In: Board R G, Jones D, Skinner F A, editors. Identification methods in applied and environmental microbiology. London, United Kingdom: Blackwell Scientific Publications Ltd.; 1992. pp. 151–161.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical