Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair - PubMed (original) (raw)
Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair
J W Hill et al. Nucleic Acids Res. 2001.
Abstract
8-Oxoguanine-DNA glycosylase 1 (OGG1), with intrinsic AP lyase activity, is the major enzyme for repairing 7,8-dihydro-8-oxoguanine (8-oxoG), a critical mutagenic DNA lesion induced by reactive oxygen species. Human OGG1 excised the damaged base from an 8-oxoG. C-containing duplex oligo with a very low apparent k(cat) of 0.1 min(-1) at 37 degrees C and cleaved abasic (AP) sites at half the rate, thus leaving abasic sites as the major product. Excision of 8-oxoG by OGG1 alone did not follow Michaelis-Menten kinetics. However, in the presence of a comparable amount of human AP endonuclease (APE1) the specific activity of OGG1 was increased approximately 5-fold and Michaelis-Menten kinetics were observed. Inactive APE1, at a higher molar ratio, and a bacterial APE (Nfo) similarly enhanced OGG1 activity. The affinity of OGG1 for its product AP.C pair (K:(d) approximately 2.8 nM) was substantially higher than for its substrate 8-oxoG.C pair (K:(d) approximately 23. 4 nM) and the affinity for its final ss-elimination product was much lower (K:(d) approximately 233 nM). These data, as well as single burst kinetics studies, indicate that the enzyme remains tightly bound to its AP product following base excision and that APE1 prevents its reassociation with its product, thus enhancing OGG1 turnover. These results suggest coordinated functions of OGG1 and APE1, and possibly other enzymes, in the DNA base excision repair pathway.
Figures
Figure 1
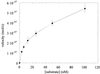
Kinetics of base excision by OGG1. OGG1 (1.25 nM) was incubated with varying concentration (3.125–100 nM) of the 8-oxoG·C-containing 32mer duplex oligo for 20 min at 37°C. Reactions were terminated by adding SDS and piperidine to 0.5% and 200 mM respectively, followed by heating at 95°C for 5 min. The products were analyzed as described in Materials and Methods.
Figure 2
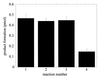
Inhibition of OGG1 glycosylase activity by AP sites. OGG1 (25 nM) was incubated with 500 nM 8-oxoG·C oligo for 5 min at 37°C and the reaction terminated as described in Figure 1. Reaction 1, OGG1 only. Identical reactions were carried out by pre-mixing with 250 nM control G·C oligo (reaction 2), β-elimination product oligo (reaction 3) or AP site-containing oligo (reaction 4).
Figure 3
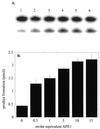
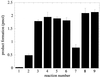
Stimulation of OGG1 glycosylase activity by APE1. OGG1 (25 nM) was incubated with the 8-oxoG·C oligo (500 nM) in the presence of varying amounts of APE1 (12.5–375 nM) and 1 mM MgCl2 for 5 min at 37°C. Termination of reactions and product analysis were performed as in Figure 1. Reaction 1, OGG1 only; reactions 2–6, OGG1 plus 12.5, 25, 125, 250 and 375 nM APE1. The actual data shown in (A) are presented as a bar graph in (B).
Figure 4
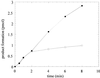
Single burst kinetics of OGG1. OGG1 (25 nM) was reacted with 500 nM 8-oxoG·C substrate for 0.5–8 min. Reactions were terminated as described in Figure 1. Open circles, OGG1 alone; closed circles, OGG1 plus a 5-fold molar excess of APE1. Data points are means of three independent experiments.
Figure 5
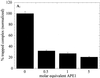
Stimulation of OGG1 glycosylase activity by mutant APE1 proteins and Nfo. OGG1 (25 nM) was incubated with 500 nM 8-oxoG·C substrate in the presence of AP endonucleases. Reactions 1 and 2, OGG1 alone. Reaction 1 was terminated with SDS and glycerol without heating. Reactions 2–9 were terminated as described in Figure 1. APE1 was added to reactions 3–8 as follows: 3, 125 nM _N_D40 APE1; 4, 125 nM _N_D60 APE1; 5, 125 nM C65S APE1; 6 and 7, 125 nM APE1; 8, 1.25 µM APE1. Reactions 1–6 and 9 contained 1 mM MgCl2. Reactions 7 and 8 contained 1 mM EDTA, reaction 9 125 nM Nfo.
Figure 6
Effect of APE1 on sodium borohydride trapping and AP lyase activity of OGG1. OGG1 (5 nM) was incubated with 1 nM 8-oxoG·C-containing oligo substrate in the presence of 1 mM sodium borohydride with varying amounts of APE1 (2.5–25 nM) for 30 min at 37°C. (A) After termination of the reaction by adding 10 µl of 2× SDS sample buffer and heating for 5 min at 100°C, the trapped complexes were separated from free substrate by SDS–PAGE. (B) Analysis of 3′-termini generated by OGG1 and APE1. Lanes 1 and 2, mobility markers for the 3′-termini characteristic of the β-elimination product and AP site, respectively. Lane 1, 1 nM 5′-labeled U·C-containing duplex oligo was incubated at 37°C for 5 min with 1 µg E.coli Udg and 1 µg Nth; lane 2, 1 nM 5′-labeled U·C oligo was incubated with 1 µg E.coli Udg and 1.25 µM APE1; lanes 3–5, 1 nM 5′-labeled 8-oxoG·C-containing oligo was incubated for 5 min at 37°C with 30 nM OGG1 (lane 3), 10 nM OGG1 and 50 nM APE1 (lane 4) or 10 nM OGG1 and 500 nM APE1 (lane 5). An aliquot of 1 mM MgCl2 was present in lanes 1–4 and 1 mM EDTA in lane 5. All reactions were terminated with SDS (0.5%) and glycerol (5%) without heating.
Figure 6
Effect of APE1 on sodium borohydride trapping and AP lyase activity of OGG1. OGG1 (5 nM) was incubated with 1 nM 8-oxoG·C-containing oligo substrate in the presence of 1 mM sodium borohydride with varying amounts of APE1 (2.5–25 nM) for 30 min at 37°C. (A) After termination of the reaction by adding 10 µl of 2× SDS sample buffer and heating for 5 min at 100°C, the trapped complexes were separated from free substrate by SDS–PAGE. (B) Analysis of 3′-termini generated by OGG1 and APE1. Lanes 1 and 2, mobility markers for the 3′-termini characteristic of the β-elimination product and AP site, respectively. Lane 1, 1 nM 5′-labeled U·C-containing duplex oligo was incubated at 37°C for 5 min with 1 µg E.coli Udg and 1 µg Nth; lane 2, 1 nM 5′-labeled U·C oligo was incubated with 1 µg E.coli Udg and 1.25 µM APE1; lanes 3–5, 1 nM 5′-labeled 8-oxoG·C-containing oligo was incubated for 5 min at 37°C with 30 nM OGG1 (lane 3), 10 nM OGG1 and 50 nM APE1 (lane 4) or 10 nM OGG1 and 500 nM APE1 (lane 5). An aliquot of 1 mM MgCl2 was present in lanes 1–4 and 1 mM EDTA in lane 5. All reactions were terminated with SDS (0.5%) and glycerol (5%) without heating.
Figure 7
Kinetics of OGG1, APE1 and Nfo. (A) OGG1 (1.25 nM) and APE1 (6.5 nM) were incubated with increasing amounts of the 8-oxoG·C-containing duplex oligo for 5 min at 37°C. (B) OGG1 (1.25 nM) was incubated with varying amounts (3.125–100 nM) of the AP·C-containing 32mer duplex oligo for 40 min at 37°C. Reactions were terminated by adding SDS and glycerol as in Figure 6B. (C) APE1 (0.5 pM) was incubated with increasing concentrations of the AP·C-containing oligo for 4 min at 37°C. (D) Escherichia coli Nfo (1.25 nM) was incubated with increasing concentrations of the AP·C-containing oligo for 4 min at 37°C. Substrates were in excess and reaction rates were in the linear range in all experiments. Data points are means of three independent experiments.
Similar articles
- Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions.
Hazra TK, Hill JW, Izumi T, Mitra S. Hazra TK, et al. Prog Nucleic Acid Res Mol Biol. 2001;68:193-205. doi: 10.1016/s0079-6603(01)68100-5. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554297 Review. - Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step.
Vidal AE, Hickson ID, Boiteux S, Radicella JP. Vidal AE, et al. Nucleic Acids Res. 2001 Mar 15;29(6):1285-92. doi: 10.1093/nar/29.6.1285. Nucleic Acids Res. 2001. PMID: 11238994 Free PMC article. - The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity.
Girard PM, Guibourt N, Boiteux S. Girard PM, et al. Nucleic Acids Res. 1997 Aug 15;25(16):3204-11. doi: 10.1093/nar/25.16.3204. Nucleic Acids Res. 1997. PMID: 9241232 Free PMC article. - Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch.
Yang H, Clendenin WM, Wong D, Demple B, Slupska MM, Chiang JH, Miller JH. Yang H, et al. Nucleic Acids Res. 2001 Feb 1;29(3):743-52. doi: 10.1093/nar/29.3.743. Nucleic Acids Res. 2001. PMID: 11160897 Free PMC article. - Mammalian Ogg1/Mmh gene plays a major role in repair of the 8-hydroxyguanine lesion in DNA.
Nishimura S. Nishimura S. Prog Nucleic Acid Res Mol Biol. 2001;68:107-23. doi: 10.1016/s0079-6603(01)68093-0. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554290 Review.
Cited by
- Modulation of base excision repair of 8-oxoguanine by the nucleotide sequence.
Allgayer J, Kitsera N, von der Lippen C, Epe B, Khobta A. Allgayer J, et al. Nucleic Acids Res. 2013 Oct;41(18):8559-71. doi: 10.1093/nar/gkt620. Epub 2013 Jul 17. Nucleic Acids Res. 2013. PMID: 23863843 Free PMC article. - In Vitro Fluorogenic Real-Time Assay of the Repair of Oxidative DNA Damage.
Edwards SK, Ono T, Wang S, Jiang W, Franzini RM, Jung JW, Chan KM, Kool ET. Edwards SK, et al. Chembiochem. 2015 Jul 27;16(11):1637-46. doi: 10.1002/cbic.201500184. Epub 2015 Jun 12. Chembiochem. 2015. PMID: 26073452 Free PMC article. - Special AT-rich Sequence-binding Protein 1 (SATB1) Functions as an Accessory Factor in Base Excision Repair.
Kaur S, Coulombe Y, Ramdzan ZM, Leduy L, Masson JY, Nepveu A. Kaur S, et al. J Biol Chem. 2016 Oct 21;291(43):22769-22780. doi: 10.1074/jbc.M116.735696. Epub 2016 Sep 2. J Biol Chem. 2016. PMID: 27590341 Free PMC article. - 17β-estradiol increases expression of the oxidative stress response and DNA repair protein apurinic endonuclease (Ape1) in the cerebral cortex of female mice following hypoxia.
Dietrich AK, Humphreys GI, Nardulli AM. Dietrich AK, et al. J Steroid Biochem Mol Biol. 2013 Nov;138:410-20. doi: 10.1016/j.jsbmb.2013.07.007. Epub 2013 Jul 29. J Steroid Biochem Mol Biol. 2013. PMID: 23907014 Free PMC article. - A unified method for purification of basic proteins.
Adhikari S, Manthena PV, Sajwan K, Kota KK, Roy R. Adhikari S, et al. Anal Biochem. 2010 May 15;400(2):203-6. doi: 10.1016/j.ab.2010.01.011. Epub 2010 Jan 28. Anal Biochem. 2010. PMID: 20109435 Free PMC article.
References
- Wiseman H., Kaur,H. and Halliwell,B. (1995) DNA damage and cancer: measurement and mechanism. Cancer Lett., 93, 113–120. - PubMed
- Beckman K.B. and Ames,B.N. (1997) Oxidative decay of DNA. J. Biol. Chem., 272, 19633–19636. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA081063/CA/NCI NIH HHS/United States
- R01 CA81063/CA/NCI NIH HHS/United States
- R01 ES008457/ES/NIEHS NIH HHS/United States
- AG10514/AG/NIA NIH HHS/United States
- R01 ES08457/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous