Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2 - PubMed (original) (raw)
Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2
Y Tao et al. Plant Cell. 2000 Dec.
Abstract
Disease resistance proteins containing a nucleotide binding site (NBS) and a leucine-rich repeat (LRR) region compose the largest class of disease resistance proteins. These so-called NBS-LRR proteins confer resistance against a wide variety of phytopathogens. To help elucidate the mechanism by which NBS-LRR proteins recognize and transmit pathogen-derived signals, we analyzed mutant versions of the Arabidopsis NBS-LRR protein RPS2. The RPS2 gene confers resistance against Pseudomonas syringae strains carrying the avirulence gene avrRpt2. The activity of RPS2 derivatives in response to AvrRpt2 was measured by using a functional transient expression assay or by expressing the mutant proteins in transgenic plants. Directed mutagenesis revealed that the NBS and an N-terminal leucine zipper (LZ) motif were critical for RPS2 function. Mutations near the N terminus, including an LZ mutation, resulted in proteins that exhibited a dominant negative effect on wild-type RPS2. Scanning the RPS2 molecule with a small in-frame internal deletion demonstrated that RPS2 does not have a large dispensable region. Overexpression of RPS2 in the transient assay in the absence of avrRpt2 also led to an apparent resistant response, presumably a consequence of a low basal activity of RPS2. The NBS and LZ were essential for this overdose effect, whereas the entire LRR was dispensable. RPS2 interaction with a 75-kD protein (p75) required an N-terminal portion of RPS2 that is smaller than the region required for the overdose effect. These findings illuminate the pathogen recognition mechanisms common among NBS-LRR proteins.
Figures
Figure 1.
RPS2 Derivatives Used in the Study. All the RPS2 derivatives used in the study are depicted in a schematic representation of the RPS2 primary structure (909 amino acid [aa] residues). The N-terminal (N-ter.) hydrophobic region, LZ, NBS motifs, LRR, and two of the motifs (GLPL and KMH) that are highly conserved among NBS-LRR proteins are shown. Amino acid substitution mutants are indicated. The lines for the C-terminal deletion mutants represent the amino acid sequence regions that remain in the mutants. The number in each C-terminal deletion mutant name represents the position of the first deleted amino acid residue. The positions of small in-frame internal deletions are indicated along the bottom line. The numbers in each in-frame internal deletion name represent the positions of the first and the last deleted amino acid residues. The mutations corresponding to deletions of <11 amino acid residues are represented by triangles; deletions of >10 amino acid residues are represented by rectangles, the widths of which correspond to the sizes of the deletions.
Figure 2.
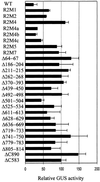
RPS2 Activity of RPS2 Derivatives Measured by a Transient Expression Assay. RPS2 activity of RPS2 derivatives in response to avrRpt2 was measured by using a transient expression assay, with a decrease in GUS activity being a measure of the RPS2 activity and with luciferase activity as a normalization factor for the transformation efficiency, as previously described (Leister et al., 1996). Similar assays were performed with RPS2 derivatives in the absence of avrRpt2. For each derivative, the data are expressed as the percentage of the normalized GUS activity observed in the presence of avrRpt2 divided by that in the absence of avrRpt2. The more active a derivative, the less its relative GUS activity. Error bars represent the standard deviation. WT, wild type.
Figure 3.
In Vivo Stability of RPS2 Derivative Proteins and Their Ability to Interact with p75. RPS2 and its derivative proteins were transiently expressed in Arabidopsis protoplasts. The RPS2 derivatives and other cellular proteins were radiolabeled with 35S-methionine, and total proteins or immunoprecipitated proteins were resolved by SDS-PAGE and visualized with a phosphorimager. (A) and (B) RPS2 and its derivatives were immunoprecipitated with an anti-RPS2 antiserum; shown are composites of the results from four independent experiments. (C) Labeled total proteins from protoplasts transfected with either RPS2 or R2M4 were precipitated with trichloroacetic acid. The amount of extract used in each lane in (C) is equivalent to ∼5% of the extract used in each lane in (A), (B), and (D). (D) A C-terminal FLAG-tagged R2M4 (left lane) and R2M4 (right lane) were immunoprecipitated with an anti-FLAG antibody and an anti-RPS2 antiserum, respectively. We attempted to normalize the results from different experiments by adjusting the imaging brightness to make comparable between experiments the intensity of the 65-kD nonspecific band (labeled by open arrowheads) for R2M4, which was included in every experiment as a positive control. The bands corresponding to the RPS2 derivatives are marked by open circles. The position of p75 is indicated by filled arrowheads. The positions of molecular mass markers are indicated at left in kilodaltons. WT, wild type.
Figure 4.
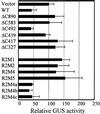
Dominant Negative Effect of R2M1 and R2M2 Measured by a Transient Expression Assay. The RPS2 wild-type construct was cobombarded with a 10-fold excess of the indicated RPS2 derivatives or the vector as competitors in a transient expression assay to measure the response to avrRpt2. The decrease in GUS activity with the vector competitor control was set to 0% inhibition, and the GUS activity observed in the absence of RPS2 (no response) was set to 100% inhibition. The error bars indicate the standard deviation.
Figure 5.
Overdose Effect of RPS2 and Its Derivatives. In the absence of the AvrRpt2 construct, we used 20-fold more of the wild-type (WT) RPS2 construct than is normally used in the transient expression assay. The same type of assay was performed with the indicated RPS2 derivatives. Relative GUS activity (normalized for transformation efficiency, as measured by luciferase activity) for the vector control was set to 100. The error bars indicate the standard deviation.
Figure 6.
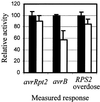
Effect of the ndr1-1 Mutation on the Transient Expression Assay. Columbia-0 (Col-0) wild-type (solid bars) and ndr1-1 plants (open bars) were tested in the transient expression assays for the response to avrRpt2, the response to avrB, and the RPS2 overdose effect. Both wild-type and ndr1-1 plants are RPS2 and RPM1 wild type. To represent the relative activity, we set the decrease of the normalized GUS activity with wild-type plants to 100, and the normalized GUS activity with vector controls was set to 0. The error bars indicate the standard deviation.
Figure 7.
Structure of pKEx4tr. The top strand of the sequence surrounding the multiple cloning site in pKEx4tr is shown. Nucleotides that differ from the corresponding nucleotides in pBI221 are shown in uppercase letters. The 35S_*_ promoter region is indicated by white letters in black background boxes. The 5′ region of the 35S_*_ promoter, which is indicated as a 761-bp region, is identical to that of the wild-type 35S promoter in pBI221. The regions derived from pUC18 are depicted schematically. The as-1 element and the TATA box of the 35S promoter, the E. coli rrnB terminator T1, some of the unique restriction sites in the vector, the T7 and T3 promoters, and the nos 3′ sequence are indicated. Plus signs indicate positions for every 10 nucleotides.
Similar articles
- Mutational analysis of the Arabidopsis RPS2 disease resistance gene and the corresponding pseudomonas syringae avrRpt2 avirulence gene.
Axtell MJ, McNellis TW, Mudgett MB, Hsu CS, Staskawicz BJ. Axtell MJ, et al. Mol Plant Microbe Interact. 2001 Feb;14(2):181-8. doi: 10.1094/MPMI.2001.14.2.181. Mol Plant Microbe Interact. 2001. PMID: 11204781 - Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1.
Leister RT, Ausubel FM, Katagiri F. Leister RT, et al. Proc Natl Acad Sci U S A. 1996 Dec 24;93(26):15497-502. doi: 10.1073/pnas.93.26.15497. Proc Natl Acad Sci U S A. 1996. PMID: 8986840 Free PMC article. - The leucine-rich repeat domain can determine effective interaction between RPS2 and other host factors in arabidopsis RPS2-mediated disease resistance.
Banerjee D, Zhang X, Bent AF. Banerjee D, et al. Genetics. 2001 May;158(1):439-50. doi: 10.1093/genetics/158.1.439. Genetics. 2001. PMID: 11333251 Free PMC article. - Use of Arabidopsis thaliana defense-related mutants to dissect the plant response to pathogens.
Ausubel FM, Katagiri F, Mindrinos M, Glazebrook J. Ausubel FM, et al. Proc Natl Acad Sci U S A. 1995 May 9;92(10):4189-96. doi: 10.1073/pnas.92.10.4189. Proc Natl Acad Sci U S A. 1995. PMID: 7753782 Free PMC article. Review. - Plant NBS-LRR proteins in pathogen sensing and host defense.
DeYoung BJ, Innes RW. DeYoung BJ, et al. Nat Immunol. 2006 Dec;7(12):1243-9. doi: 10.1038/ni1410. Nat Immunol. 2006. PMID: 17110940 Free PMC article. Review.
Cited by
- RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance.
Bieri S, Mauch S, Shen QH, Peart J, Devoto A, Casais C, Ceron F, Schulze S, Steinbiss HH, Shirasu K, Schulze-Lefert P. Bieri S, et al. Plant Cell. 2004 Dec;16(12):3480-95. doi: 10.1105/tpc.104.026682. Epub 2004 Nov 17. Plant Cell. 2004. PMID: 15548741 Free PMC article. - Ectopic Expression in Arabidopsis thaliana of an NB-ARC Encoding Putative Disease Resistance Gene from Wild Chinese Vitis pseudoreticulata Enhances Resistance to Phytopathogenic Fungi and Bacteria.
Wen Z, Yao L, Wan R, Li Z, Liu C, Wang X. Wen Z, et al. Front Plant Sci. 2015 Dec 10;6:1087. doi: 10.3389/fpls.2015.01087. eCollection 2015. Front Plant Sci. 2015. PMID: 26697041 Free PMC article. - Full-genome analysis of resistance gene homologues in rice.
Monosi B, Wisser RJ, Pennill L, Hulbert SH. Monosi B, et al. Theor Appl Genet. 2004 Nov;109(7):1434-47. doi: 10.1007/s00122-004-1758-x. Epub 2004 Aug 10. Theor Appl Genet. 2004. PMID: 15309302 - Identification and mapping of a novel dominant resistance gene, TuRB07 to Turnip mosaic virus in Brassica rapa.
Jin M, Lee SS, Ke L, Kim JS, Seo MS, Sohn SH, Park BS, Bonnema G. Jin M, et al. Theor Appl Genet. 2014 Feb;127(2):509-19. doi: 10.1007/s00122-013-2237-z. Epub 2013 Dec 18. Theor Appl Genet. 2014. PMID: 24346479 - Meristem micropropagation of cassava (Manihot esculenta) evokes genome-wide changes in DNA methylation.
Kitimu SR, Taylor J, March TJ, Tairo F, Wilkinson MJ, Rodríguez López CM. Kitimu SR, et al. Front Plant Sci. 2015 Aug 13;6:590. doi: 10.3389/fpls.2015.00590. eCollection 2015. Front Plant Sci. 2015. PMID: 26322052 Free PMC article.
References
- Assaad, F.F., and Signer, E.R. (1990). Cauliflower mosaic virus P35S promoter activity in Escherichia coli. Mol. Gen. Genet. 223 517–520. - PubMed
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1998). Current Protocols in Molecular Biology. (New York: John Wiley and Sons).
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265 1856–1860. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous