The phagosome proteome: insight into phagosome functions - PubMed (original) (raw)
The phagosome proteome: insight into phagosome functions
J Garin et al. J Cell Biol. 2001.
Abstract
Phagosomes are key organelles for the innate ability of macrophages to participate in tissue remodeling, clear apoptotic cells, and restrict the spread of intracellular pathogens. To understand the functions of phagosomes, we initiated the systematic identification of their proteins. Using a proteomic approach, we identified >140 proteins associated with latex bead-containing phagosomes. Among these were hydrolases, proton pump ATPase subunits, and proteins of the fusion machinery, validating our approach. A series of unexpected proteins not previously described along the endocytic/phagocytic pathways were also identified, including the apoptotic proteins galectin3, Alix, and TRAIL, the anti-apoptotic protein 14-3-3, the lipid raft-enriched flotillin-1, the anti-microbial molecule lactadherin, and the small GTPase rab14. In addition, 24 spots from which the peptide masses could not be matched to entries in any database potentially represent new phagosomal proteins. The elaboration of a two-dimensional gel database of >160 identified spots allowed us to analyze how phagosome composition is modulated during phagolysosome biogenesis. Remarkably, during this process, hydrolases are not delivered in bulk to phagosomes, but are instead acquired sequentially. The systematic characterization of phagosome proteins provided new insights into phagosome functions and the protein or groups of proteins involved in and regulating these functions.
Figures
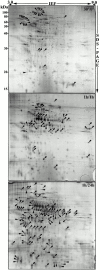
Figure 1
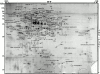
Phagosome protein 2-D gel map. Latex beads were internalized by J774 macrophages for 60 min followed by a 60-min incubation without bead. After cell breakage, phagosomes were isolated on sucrose gradients and their proteins separated by high-resolution 2-D gel electrophoresis. Proteins were separated according to their isoelectric point on immobilized pH-gradients 3-10, and then by standard SDS-PAGE. The major spots were excised and analyzed by mass spectrometry. The image used here is a representative gel stained with silver.
Figure 2
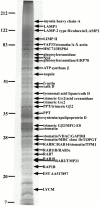
Isolation of phagosome membrane-associated proteins by Triton X-114 extraction. Latex beads were internalized by J774 macrophages for 60 min followed by a 60-min incubation without bead. Isolated phagosomes were then treated with Triton X-114 to separate membrane-associated proteins from soluble proteins. Proteins present in the detergent phase were then separated by SDS-PAGE. The gels were stained with zinc acetate and the major bands excised for mass spectrometry analysis. In some cases, several proteins were identified by mass fingerprinting in the same band.
Figure 3
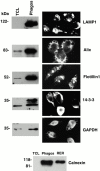
Immunolocalization of novel proteins to phagosomes. To demonstrate the phagosomal association of the new proteins identified in our study, we performed Western blot and immunofluorescence analyses of some of these proteins in macrophages that had internalized latex beads for 60 min, followed by a 60-min chase. LAMP1, a well known phagosomal protein, was used as a control. For Western blot analysis, the same amount of protein from total cell lysates or isolated phagosomes was loaded on SDS-PAGE. A clear enrichment of each protein on the phagosomes is observed. Immunofluorescence analysis clearly demonstrate the localization of the protein around compartments containing latex beads. In the case of 14-3-3 and GAPDH, cells were permeabilized before the labeling to get rid of most of the cytosolic proteins. Note that one of the cells has not been permeabilized in the case of 14-3-3. For calnexin, an antibody against the cytosolic portion of the molecule was used. The same amount of protein was loaded for total cell lysate (TCL), the phagosomes (Phagos), and a microsome preparation (ER) as a positive control.
Figure 4
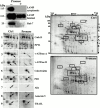
Sensitivity of phagosome proteins to protease treatment: lumenal versus cytoplasmic proteins. (A) Phagosomes isolated on sucrose gradients were treated with pronase to digest proteins exposed on the cytoplasmic side of phagosomes. Western blot analyses showed that the lumenal part of LAMP1, a transmembrane protein with a short cytoplasmic tail, was not affected by the pronase treatment, while the cytoplasmic tail was degraded. Rab7, a protein associated with the cytoplasmic side of phagosomes was efficiently degraded (or released) by the treatment. (B) Comparison of the protein patterns between phagosomes treated or not with pronase shows that several protein spots are no longer present on phagosomes after the pronase treatment (arrows), indicating they have been digested by the proteases. (C) Insets of selected regions of the gel show that hydrolases present within the lumen of phagosomes, such as cathepsins A and D, are not affected by the pronase treatment. In contrast, proteins like the A and B subunits of the vacuolar proton pump, or proteins associated with cytoskeletal elements, which are exposed on the cytoplasmic side of phagosomes, are degraded and almost absent on the 2-D gel. Interestingly, proteins of the ER, such as calreticulin and PDI, are not degraded, indicating that these ER elements are within the lumen of phagosomes and are thus unlikely to constitute contaminants trapped by cytoskeletal elements on the outside of phagosomes, which would be released by the treatment. Another interesting observation is the presence of TRAIL and Annexin5 in the lumen of the phagosome, in contrast to Alix on the cytoplasmic side, as expected from proteins involved in apoptotic signaling.
Figure 5
Modulation of phagosome composition during maturation. Latex bead–containing phagosomes were isolated at different stages of maturation and their proteins separated by 2-D gel electrophoresis. The various gels were then compared with each other using the software PD-Quest. Arrows point to peaking proteins at their respective time points.
Figure 6
Sequential acquisition of hydrolases during phagosome maturation. Phagosomes were formed by the internalization of latex beads for 30 or 60 min, followed by increasing periods of chase to allow phagosome maturation. After 2-D gel electrophoresis, the protein patterns were analyzed using the phagosome 2-D database (Fig. 1). These analyses indicated that hydrolases are not acquired simultaneously by phagosomes, but rather appear sequentially during phagolysosome biogenesis.
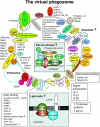
Figure 7
The virtual phagosome. The major proteins identified in the present study are presented in their potential interaction with phagosomes. In several cases, the localization to the lumen, the membrane, or the cytoplasmic aspect of the phagosome was indicated by the sensitivity to pronase proteolysis (Fig. 4). (Insets) New concepts proposed after the identification of novel phagosome proteins.
Similar articles
- The Mycobacterium bovis bacille Calmette-Guerin phagosome proteome.
Lee BY, Jethwaney D, Schilling B, Clemens DL, Gibson BW, Horwitz MA. Lee BY, et al. Mol Cell Proteomics. 2010 Jan;9(1):32-53. doi: 10.1074/mcp.M900396-MCP200. Epub 2009 Oct 7. Mol Cell Proteomics. 2010. PMID: 19815536 Free PMC article. - Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes.
Dermine JF, Duclos S, Garin J, St-Louis F, Rea S, Parton RG, Desjardins M. Dermine JF, et al. J Biol Chem. 2001 May 25;276(21):18507-12. doi: 10.1074/jbc.M101113200. Epub 2001 Feb 27. J Biol Chem. 2001. PMID: 11279173 - Proteomic analysis reveals a role for protein kinase C-alpha in phagosome maturation.
Ng Yan Hing JD, Desjardins M, Descoteaux A. Ng Yan Hing JD, et al. Biochem Biophys Res Commun. 2004 Jul 2;319(3):810-6. doi: 10.1016/j.bbrc.2004.05.054. Biochem Biophys Res Commun. 2004. PMID: 15184055 - Regulators of membrane trafficking and Mycobacterium tuberculosis phagosome maturation block.
Fratti RA, Vergne I, Chua J, Skidmore J, Deretic V. Fratti RA, et al. Electrophoresis. 2000 Oct;21(16):3378-85. doi: 10.1002/1522-2683(20001001)21:16<3378::AID-ELPS3378>3.0.CO;2-B. Electrophoresis. 2000. PMID: 11079558 Review. - Analysis of phagosomal proteomes: from latex-bead to bacterial phagosomes.
Li Q, Jagannath C, Rao PK, Singh CR, Lostumbo G. Li Q, et al. Proteomics. 2010 Nov;10(22):4098-116. doi: 10.1002/pmic.201000210. Proteomics. 2010. PMID: 21080496 Free PMC article. Review.
Cited by
- Protein disulfide isomerase and host-pathogen interaction.
Stolf BS, Smyrnias I, Lopes LR, Vendramin A, Goto H, Laurindo FR, Shah AM, Santos CX. Stolf BS, et al. ScientificWorldJournal. 2011;11:1749-61. doi: 10.1100/2011/289182. Epub 2011 Oct 18. ScientificWorldJournal. 2011. PMID: 22125433 Free PMC article. Review. - Host-Pathogen Interactions of Marine Gram-Positive Bacteria.
Gnanagobal H, Santander J. Gnanagobal H, et al. Biology (Basel). 2022 Sep 5;11(9):1316. doi: 10.3390/biology11091316. Biology (Basel). 2022. PMID: 36138795 Free PMC article. Review. - The Leishmania donovani lipophosphoglycan excludes the vesicular proton-ATPase from phagosomes by impairing the recruitment of synaptotagmin V.
Vinet AF, Fukuda M, Turco SJ, Descoteaux A. Vinet AF, et al. PLoS Pathog. 2009 Oct;5(10):e1000628. doi: 10.1371/journal.ppat.1000628. Epub 2009 Oct 16. PLoS Pathog. 2009. PMID: 19834555 Free PMC article. - Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis.
Müller-Taubenberger A, Lupas AN, Li H, Ecke M, Simmeth E, Gerisch G. Müller-Taubenberger A, et al. EMBO J. 2001 Dec 3;20(23):6772-82. doi: 10.1093/emboj/20.23.6772. EMBO J. 2001. PMID: 11726513 Free PMC article. - The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins.
Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. Carter C, et al. Plant Cell. 2004 Dec;16(12):3285-303. doi: 10.1105/tpc.104.027078. Epub 2004 Nov 11. Plant Cell. 2004. PMID: 15539469 Free PMC article.
References
- Alvarez-Dominguez C., Stahl P.D. Increased expression of Rab5a correlates directly with accelerated maturation of Listeria monocytogenes phagosomes. J. Biol. Chem. 1999;274:11459–11462. - PubMed
- Berón W., Colombo M.I., Mayorga L.S., Stahl P.D. In vitro reconstitution of phagosome-endosome fusionevidence for regulation by heterotrimeric GTPases. Arch. Biochem. Biophys. 1995;317:337–342. - PubMed
- Bickel P.E., Scherer P.E., Schnitzer J.E., Oh P., Lisanti M.P., Lodish H.F. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J. Biol. Chem. 1997;272:13793–13802. - PubMed
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 1981;256:1604–1607. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous