An evolutionarily conserved transmembrane protein that is a novel downstream target of neurotrophin and ephrin receptors - PubMed (original) (raw)
An evolutionarily conserved transmembrane protein that is a novel downstream target of neurotrophin and ephrin receptors
H Kong et al. J Neurosci. 2001.
Abstract
Appropriate development of nervous system connectivity involves a variety of processes, including neuronal life-and-death decisions, differentiation, axon guidance and migration, and synaptogenesis. Although these activities likely require specialized signaling events, few substrates unique to these neurotrophic functions have been identified. Here we describe the cloning of ankyrin repeat-rich membrane spanning (ARMS), which encodes a novel downstream target of neurotrophin and ephrin receptor tyrosine kinases, Trk and Eph, respectively. The amino acid sequence of ARMS is highly conserved from nematode to human, suggesting an evolutionarily conserved role for this protein. The ARMS protein consists of 1715 amino acids containing four putative transmembrane domains, multiple ankyrin repeats, a sterile alpha motif domain, and a potential PDZ-binding motif. In the rat, ARMS is specifically expressed in the developing nervous system and in highly plastic areas of the adult brain, regions enriched in Trks and Eph receptors. ARMS can physically associate with TrkA and p75 neurotrophin receptors. Moreover, endogenous ARMS protein is tyrosine phosphorylated after neurotrophin treatment of pheochromocytoma 12 cells and primary hippocampal neurons or ephrin B treatment of NG108-15 cells, demonstrating that ARMS is a downstream target for both neurotrophin and ephrin receptors.
Figures
Fig. 1.
ARMS represents a novel transmembrane protein.A, Predicted topology of ARMS. Transmembrane domains (labeled 1–4) and various intracellular motifs are depicted.B, Amino acid sequence and comparison of rat ARMS (rARMS) and human ARMS (hARMS) proteins. Residues marked with a_dashed line_ denote 11 contiguous ankyrin repeats;bold-faced tyrosine (Y) residues (at positions 399, 409, 441, 444, and 466 of the rat sequence) are evolutionarily conserved among human, rat, Drosophila, and C. elegans; boxed residues are the putative transmembrane domains; italicized residues denote the polyproline stretch; shadowed residues constitute the SAM domain (amino acids 1152–1221); and the three carboxymost amino acids marked with asterisks (SIL) encode a potential PDZ-binding motif. Amino acids of the human sequence that differ from the rat sequence are shown. C, Comparison of various cytoplasmic regions of rat (r), human (h), Drosophila(d), and C. elegans(w) ARMS proteins. Part 1, N-terminal region between the ankyrin repeats and the first transmembrane domain with bold-faced evolutionarily conserved tyrosines (Y). Part 2, Cytoplasmic region between transmembrane domains 2 and 3. Parts 3, 4, Two C-terminal regions. Part 5, The SAM domain. Sequences for wARMS, dARMS, and hARMS were obtained from accession numbersZ68760, AAF46710, and BAA86564/CAB63746, respectively.Symbols: asterisk, identity;colon, strongly similar; period, weakly similar.
Fig. 1.
ARMS represents a novel transmembrane protein.A, Predicted topology of ARMS. Transmembrane domains (labeled 1–4) and various intracellular motifs are depicted.B, Amino acid sequence and comparison of rat ARMS (rARMS) and human ARMS (hARMS) proteins. Residues marked with a_dashed line_ denote 11 contiguous ankyrin repeats;bold-faced tyrosine (Y) residues (at positions 399, 409, 441, 444, and 466 of the rat sequence) are evolutionarily conserved among human, rat, Drosophila, and C. elegans; boxed residues are the putative transmembrane domains; italicized residues denote the polyproline stretch; shadowed residues constitute the SAM domain (amino acids 1152–1221); and the three carboxymost amino acids marked with asterisks (SIL) encode a potential PDZ-binding motif. Amino acids of the human sequence that differ from the rat sequence are shown. C, Comparison of various cytoplasmic regions of rat (r), human (h), Drosophila(d), and C. elegans(w) ARMS proteins. Part 1, N-terminal region between the ankyrin repeats and the first transmembrane domain with bold-faced evolutionarily conserved tyrosines (Y). Part 2, Cytoplasmic region between transmembrane domains 2 and 3. Parts 3, 4, Two C-terminal regions. Part 5, The SAM domain. Sequences for wARMS, dARMS, and hARMS were obtained from accession numbersZ68760, AAF46710, and BAA86564/CAB63746, respectively.Symbols: asterisk, identity;colon, strongly similar; period, weakly similar.
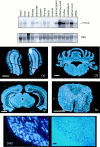
Fig. 2.
Distribution of ARMS mRNA. Top, Northern analysis of ARMS. A single transcript of 7.0 kb was detected by Northern analysis using a 32P-labeled ARMS cDNA probe (top blot). Each lane contained 20 μg of total RNA (with the exception of the pancreas and DRG_lanes_ that contained <10 μg each) extracted from various rat tissue. Methylene blue staining of the 28 S ribosomal band as a loading control is shown (bottom blot).Middle, Distribution of ARMS mRNA in the adult rat CNS. A 33P-labeled cRNA probe was used to assess ARMS mRNA expression. Areas of intense labeling include the mitral cell layer of the olfactory bulb (OB), all regions of the hippocampus (HP), the Purkinje cell layer of the cerebellum (CB), and gray matter (most notably in the ventral horn) of the spinal cord (SC). Bottom, Expression in adult rat DRG by in situ hybridization. A33P-labeled cRNA probe was used to assess mRNA distribution in DRG as depicted in the dark-field image (left). The majority of cell bodies of the DRG were positive for ARMS mRNA expression, but absences of expression were noted in the large-diameter DRG cell bodies depicted by the arrows in the dark-field and the corresponding phase (right) photographs. Scale bars: white, 1 mm; black, 50 μm.
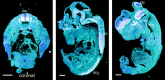
Fig. 3.
Expression of ARMS mRNA. _In situ_hybridization of ARMS in E14 rat. Left, In a coronal section through the midsection of an E14 rat, only spinal cord (sc) and dorsal root ganglion (drg) were positive for ARMS. Middle, Right,In situ hybridization of ARMS in a midsagittal section and a more lateral section, respectively, of E14 rat is shown. ARMS mRNA expression was restricted to various brain regions such as the cortex (cx), hippocampus (hp), pons, medulla (med), basal telencephalon (bt), principal and spinal trigeminal nucleus (tn), superior and inferior colliculus (clc), and sc. Multiple ganglia expressed ARMS mRNA, such as the drg, trigeminal ganglion (tg), geniculate ganglion (gg), vestibular ganglion (vg), and superior cervical ganglion (scg). Scale bars, 1 mm.
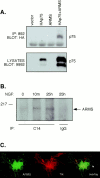
Fig. 4.
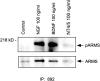
Association of ARMS with p75 and TrkA receptors. A, Interaction of p75 with ARMS. HEK293T cells were cotransfected with cDNAs encoding full-length ARMS (ARMS), HA-tagged p75 (HAp75), ARMS plus p75 (HAp75+ARMS), or empty vector (vector). Cells lysates were immunoprecipitated with anti-ARMS 892 antiserum and immunoblotted with anti-HA (top). Expression of p75 receptors was confirmed by immunoblotting with anti-p75 (9992; bottom).B, Coprecipitation of TrkA and ARMS. PC12 615 cells were treated for 10 min (m) and 25 hr (h) with NGF (100 ng/ml). Lysates were prepared and subjected to immunoprecipitation with anti-Trk C14 antibody, followed by immunoblotting with anti-ARMS antibody. Normal rabbit IgG was used as a negative control. The migration of the protein molecular weight standard, 217 kDa, is shown on the left. C, Colocalization of ARMS and TrkA. Immunofluorescence analysis of ARMS and TrkA receptor in sympathetic neurons is shown. SCG sympathetic neurons were grown in the presence of 150 ng/ml NGF, fixed, and immunostained as described in Materials and Methods. The ARMS protein and the TrkA receptor were subjected to double immunostaining using an anti-ARMS antiserum (left) and an anti-Trk B-3 monoclonal antibody (middle) and were analyzed by confocal microscopy. The yellow signal demonstrates overlap of the two signals (overlay; right). The_arrow_ indicates cell surface colocalization of ARMS and TrkA. IP, Immunoprecipitation.
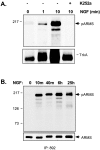
Fig. 5.
Tyrosine phosphorylation of ARMS.A, Phosphorylation of ARMS by NGF in PC12 cells is rapid and can be blocked by K252a. The antiserum 892 was used to immunoprecipitate endogenously expressed ARMS from PC12 615 cell lysates. Anti-phosphotyrosine antibody pY99 was used to assess tyrosine phosphorylation of the immunoprecipitated ARMS. Within 1 min of NGF treatment, phosphorylated ARMS (pARMS) could be detected, suggesting a direct phosphorylation by TrkA. Furthermore, 100 n
m
K252a potently blocked ARMS phosphorylation (top). In lysates of the same samples, TrkA autophosphorylation is shown using pY99 (bottom).B, Time course of ARMS phosphorylation by NGF in PC12 cells is shown. The phosphorylation peaks within 10 min and is sustained for at least 25 hr (top). Reprobing of the same blot with 892 demonstrated equivalent levels of immunoprecipitated ARMS from the various lysates (bottom).
Fig. 6.
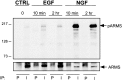
Specificity of ARMS phosphorylation.Top, Phosphorylation of ARMS is specifically induced after NGF, but not EGF, treatment of PC12 615 cells. Two time points, 10 min and 2 hr, were examined for tyrosine phosphorylation of ARMS using the following conditions: no ligand (CTRL), 50 ng/ml EGF, and 100 ng/ml NGF. To demonstrate the specificity of the ARMS antiserum 892 (I), preimmune antiserum (P) was used in parallel immunoprecipitations. Bottom, The amount of ARMS protein that was immunoprecipitated from the various lysates is shown.
Fig. 7.
Phosphorylation of ARMS by other neurotrophins. The neurotrophins BDNF and NT-4/5 induce phosphorylation of ARMS via the TrkB receptor. PC12 cells stably expressing TrkB were treated with either 100 ng/ml BDNF or 100 ng/ml NT-4/5, and the phosphorylation of ARMS was measured as described in Figure 5. Top, BDNF and, to a lesser extent, NT-4/5 were able to induce tyrosine phosphorylation of ARMS. Bottom, Immunoprecipitated ARMS from each lysate is shown.
Fig. 8.
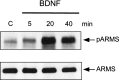
Induction of ARMS phosphorylation in hippocampal neurons by BDNF. Primary cultures of E17 hippocampal neurons were prepared and treated with 50 ng/ml BDNF for the indicated times.Top, Phosphorylation of ARMS was assessed by immunoprecipitation with anti-ARMS 892 antiserum and Western blotting with anti-phosphotyrosine pY99 antibody. Bottom, Equal amounts of ARMS protein were immunoprecipitated from each lysate as shown by reprobing the same blot with 892. C, Control.
Fig. 9.
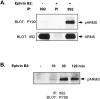
Phosphorylation of ARMS by ephrins.A, Ephrin B2 induces ARMS tyrosine phosphorylation in NG108-15 cells expressing EphB2 receptor. Lysates were made from untreated or ligand-stimulated NG108-15 cells (using aggregated ephrin B2; 30–40 min) and immunoprecipitated with 892 antiserum or preimmune (PI) serum. Tyrosine phosphorylation was assessed with pY99 in subsequent Western blots (top). Equivalent amounts of ARMS were immunoprecipitated (bottom).B, Tyrosine phosphorylation of ARMS by ephrin B2 peaks at 30 min. Thus, the time course of ARMS tyrosine phosphorylation closely parallels that of receptor autophosphorylation.
Similar articles
- Structural determinants of Trk receptor specificities using BDNF-based neurotrophin chimeras.
Lai KO, Glass DJ, Geis D, Yancopoulos GD, Ip NY. Lai KO, et al. J Neurosci Res. 1996 Dec 1;46(5):618-29. doi: 10.1002/(SICI)1097-4547(19961201)46:5<618::AID-JNR10>3.0.CO;2-T. J Neurosci Res. 1996. PMID: 8951673 - Identification of a switch in neurotrophin signaling by selective tyrosine phosphorylation.
Arévalo JC, Pereira DB, Yano H, Teng KK, Chao MV. Arévalo JC, et al. J Biol Chem. 2006 Jan 13;281(2):1001-7. doi: 10.1074/jbc.M504163200. Epub 2005 Nov 11. J Biol Chem. 2006. PMID: 16284401 - p75 reduces TrkB tyrosine autophosphorylation in response to brain-derived neurotrophic factor and neurotrophin 4/5.
Vesa J, Kruttgen A, Shooter EM. Vesa J, et al. J Biol Chem. 2000 Aug 11;275(32):24414-20. doi: 10.1074/jbc.M001641200. J Biol Chem. 2000. PMID: 10825163 - Molecular interactions between neurotrophin receptors.
Dechant G. Dechant G. Cell Tissue Res. 2001 Aug;305(2):229-38. doi: 10.1007/s004410100378. Cell Tissue Res. 2001. PMID: 11545260 Review. - p75 and Trk: a two-receptor system.
Chao MV, Hempstead BL. Chao MV, et al. Trends Neurosci. 1995 Jul;18(7):321-6. Trends Neurosci. 1995. PMID: 7571013 Review.
Cited by
- Upregulated ankyrin repeat-rich membrane spanning protein contributes to tumour progression in cutaneous melanoma.
Liao YH, Hsu SM, Yang HL, Tsai MS, Huang PH. Liao YH, et al. Br J Cancer. 2011 Mar 15;104(6):982-8. doi: 10.1038/bjc.2011.18. Epub 2011 Feb 22. Br J Cancer. 2011. PMID: 21343931 Free PMC article. - Early structural and functional plasticity alterations in a susceptibility period of DYT1 dystonia mouse striatum.
Maltese M, Stanic J, Tassone A, Sciamanna G, Ponterio G, Vanni V, Martella G, Imbriani P, Bonsi P, Mercuri NB, Gardoni F, Pisani A. Maltese M, et al. Elife. 2018 Mar 5;7:e33331. doi: 10.7554/eLife.33331. Elife. 2018. PMID: 29504938 Free PMC article. - In vivo functions of p75NTR: challenges and opportunities for an emerging therapeutic target.
Malik SC, Sozmen EG, Baeza-Raja B, Le Moan N, Akassoglou K, Schachtrup C. Malik SC, et al. Trends Pharmacol Sci. 2021 Sep;42(9):772-788. doi: 10.1016/j.tips.2021.06.006. Epub 2021 Jul 29. Trends Pharmacol Sci. 2021. PMID: 34334250 Free PMC article. Review. - Bex3 Dimerization Regulates NGF-Dependent Neuronal Survival and Differentiation by Enhancing trkA Gene Transcription.
Calvo L, Anta B, López-Benito S, Martín-Rodriguez C, Lee FS, Pérez P, Martín-Zanca D, Arévalo JC. Calvo L, et al. J Neurosci. 2015 May 6;35(18):7190-202. doi: 10.1523/JNEUROSCI.4646-14.2015. J Neurosci. 2015. PMID: 25948268 Free PMC article. - Transient Receptor Potential Vanilloid 1 Signaling Is Independent on Protein Kinase A Phosphorylation of Ankyrin-Rich Membrane Spanning Protein.
Pellegrino A, Mükusch S, Seitz V, Stein C, Herberg FW, Seitz H. Pellegrino A, et al. Med Sci (Basel). 2022 Nov 17;10(4):63. doi: 10.3390/medsci10040063. Med Sci (Basel). 2022. PMID: 36412904 Free PMC article.
References
- Aibel L, Martin-Zanca D, Perez P, Chao MV. Functional expression of TrkA receptors in hippocampal neurons. J Neurosci Res. 1998;54:424–431. - PubMed
- Barker PA, Shooter EM. Disruption of NGF binding to the low affinity neurotrophin receptor p75LNTR reduces NGF binding to TrkA on PC12 cells. Brain Res. 1994;664:155–166. - PubMed
- Berg MM, Sternberg DW, Parada LF, Chao MV. K-252a inhibits nerve growth factor-induced trk proto-oncogene tyrosine phosphorylation and kinase activity. J Biol Chem. 1992;267:13–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HD 23315/HD/NICHD NIH HHS/United States
- NS 21072/NS/NINDS NIH HHS/United States
- P50 DA005010/DA/NIDA NIH HHS/United States
- P01 HD023315/HD/NICHD NIH HHS/United States
- R01 NS021072/NS/NINDS NIH HHS/United States
- F32 NS010489/NS/NINDS NIH HHS/United States
- NS 10489/NS/NINDS NIH HHS/United States
- R56 NS021072/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous