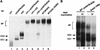Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56 - PubMed (original) (raw)
Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56
M Zhang et al. Genes Dev. 2001.
Abstract
The human 56-kD U2AF(65)-associated protein (hUAP56), a member of the DExD/H box protein family of RNA-dependent ATPases, is required for the stable binding of U2 snRNP to the pre-mRNA branchpoint. Here we identify a highly conserved Saccharomyces cerevisiae homolog of hUAP56, yUAP/Sub2p. yUAP/Sub2p can be functionally substituted for by hUAP56 and, like its human counterpart, is an essential pre-mRNA splicing factor. yUAP/Sub2p is required for formation of the commitment complex, the precursor for U2 snRNP addition. In conjunction with previous studies, we conclude that at least two DExD/H box proteins, Prp5p and yUAP/Sub2p, mediate the U2 snRNP-branchpoint interaction.
Figures
Figure 1
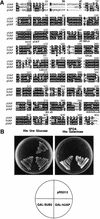
A highly conserved Saccharomyces cerevisiae homolog of hUAP56. (A) Amino acid alignment. The S. cerevisiae yUAP/Sub2p amino acid sequence was aligned to human UAP56 (hUAP) and the hypothetical Schizosaccharomyces pombe UAP (pUAP) found in database searches. Identical amino acids in all three homologs are shaded. Conserved motifs characteristic of DExD/H box protein family are indicated at the bottom of the alignment. Star indicates the mutated aspartic acid residue in sub2-100, a temperature-sensitive allele of SUB2. (B) hUAP56 can functionally substitute for yUAP/Sub2p. Growth of cells on 5FOA, galactose plate is shown.
Figure 2
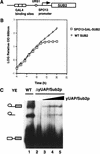
yUAP/Sub2p is an essential pre-mRNA splicing factor. (A) Schematic diagram of a conditional yUAP/Sub2p expression construct. (B) Growth curve of strain MZ105, which conditionally expresses yUAP/Sub2p. Strain MZ102a, which expresses yUAP/Sub2p constitutively, was used as a control. Cultures of MZ105 and MZ102a in 4% galactose containing medium were centrifuged and the cells resuspended in rich medium containing 4% glucose at time zero. Every 2 h, the OD600 of the cultures was measured and the concentration of cells adjusted to remain between OD600 1.0 and 3.0. (C) Splicing analysis. Splicing reactions were carried out at 25°C for 30 min. In lane 1, splicing extract was prepared from strain MZ105 grown in galactose-containing media. In lanes 2_–_5, splicing extract was prepared from strain MZ105 16 h following shift from galactose- to glucose-containing media. In lanes 3_–_5, 0.1 μL, 0.5 μ, and 1 μL of GST-yUAP/Sub2p (0.1 mg/mL) was added to the reaction mixture.
Figure 3
A temperature-sensitive mutant of yUAP/Sub2p. (A) Growth phenotype of sub2-100 containing strain, MZ306. (WT yUAP/Sub2), yeast strain MZ102b carrying SUB2. (Sub2-100), yeast strain MZ306 carrying a mutant allele of _SUB2, sub2_–100. (B) Splicing analysis. Splicing extracts were derived from strain MZ102b (lanes 1,2) and MZ306 grown at 30°C (lanes 3,4,5), respectively. Splicing reactions were carried out at 25°C for 30 min. In lane 4, the splicing extract was preincubated at 37°C for 5 min. In lanes 2 and 5, splicing extracts were preincubated at 37°C for 15 min.
Figure 4
yUAP/Sub2p is required for commitment complex formation. (A) Analysis of splicing complex formation using U1 snRNA-, U2 snRNA-, or yUAP/Sub2p-depleted extracts. yUAP/Sub2p-, U1 snRNA-, or U2 snRNA-depleted splicing extract was derived from strain MZ105, BS-Y82, or BS-Y88, respectively, 16 h after shift from galactose- to glucose-containing media. Splicing reactions (40% extract) were carried out and analyzed on a native gel. In lanes 4–6, 20% of the indicated extracts were used. In lanes 7 and 8, extracts were prepared from strain W303 and MZ105 grown in galactose-containing media. Commitment (CC1 and CC2) and spliceosomal (SP) complexes are indicated. (B) Analysis of splicing complex formation using sub2-100 containing extracts. Splicing extracts were derived from strain MZ102b (lanes 1,2), and MZ306 grown at 30°C (lanes 3,4), respectively. Splicing reactions (40% extract) were carried out for 20 min and analyzed on a native gel. In lanes 2 and 4, splicing extracts were preincubated at 37°C for 15 min before assembling the splicing reaction mixture.
Figure 5
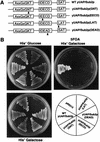
Role of DExD/H box protein family consensus motifs in yUAP/Sub2p function. (A) Amino acid sequences of the mutated motifs. Arrows indicate the mutated residues. (B) Effects of yUAP/Sub2p mutations on cell growth. Strains used in top panel were derived from strain MZ102b and contain plasmid pMZ11 (SUB2, URA3, CEN/ARS) and an integrated copy of wild-type yUAP/Sub2p (MZ104) or various yUAP/Sub2p mutants (MZ106, MZ107, MZ108, and MZ109) expressed from the G_AL1_ promoter. Growth of cells carrying the indicated genes on His− glucose or 5FOA His− galactose is shown. In the bottom panel, W303 was used as the parental strain. Growth of cells on a His− galactose plate is shown.
Similar articles
- U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction.
Fleckner J, Zhang M, Valcárcel J, Green MR. Fleckner J, et al. Genes Dev. 1997 Jul 15;11(14):1864-72. doi: 10.1101/gad.11.14.1864. Genes Dev. 1997. PMID: 9242493 - The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition.
Abovich N, Liao XC, Rosbash M. Abovich N, et al. Genes Dev. 1994 Apr 1;8(7):843-54. doi: 10.1101/gad.8.7.843. Genes Dev. 1994. PMID: 7926772 - Identification and characterization of a yeast gene encoding the U2 small nuclear ribonucleoprotein particle B" protein.
Tang J, Abovich N, Rosbash M. Tang J, et al. Mol Cell Biol. 1996 Jun;16(6):2787-95. doi: 10.1128/MCB.16.6.2787. Mol Cell Biol. 1996. PMID: 8649387 Free PMC article. - Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase.
Kistler AL, Guthrie C. Kistler AL, et al. Genes Dev. 2001 Jan 1;15(1):42-9. doi: 10.1101/gad.851301. Genes Dev. 2001. PMID: 11156604 Free PMC article. - mRNA export: travelling with DEAD box proteins.
Linder P, Stutz F. Linder P, et al. Curr Biol. 2001 Nov 27;11(23):R961-3. doi: 10.1016/s0960-9822(01)00574-7. Curr Biol. 2001. PMID: 11728322 Review.
Cited by
- The Hog1 MAPK substrate governs Candida glabrata-epithelial cell adhesion via the histone H2A variant.
Sahu MS, Purushotham R, Kaur R. Sahu MS, et al. PLoS Genet. 2024 May 14;20(5):e1011281. doi: 10.1371/journal.pgen.1011281. eCollection 2024 May. PLoS Genet. 2024. PMID: 38743788 Free PMC article. - Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA.
Preker PJ, Guthrie C. Preker PJ, et al. RNA. 2006 Jun;12(6):994-1006. doi: 10.1261/rna.6706. Epub 2006 Apr 17. RNA. 2006. PMID: 16618971 Free PMC article. - Cus2 enforces the first ATP-dependent step of splicing by binding to yeast SF3b1 through a UHM-ULM interaction.
Talkish J, Igel H, Hunter O, Horner SW, Jeffery NN, Leach JR, Jenkins JL, Kielkopf CL, Ares M. Talkish J, et al. RNA. 2019 Aug;25(8):1020-1037. doi: 10.1261/rna.070649.119. Epub 2019 May 20. RNA. 2019. PMID: 31110137 Free PMC article. - Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism.
Preker PJ, Kim KS, Guthrie C. Preker PJ, et al. RNA. 2002 Aug;8(8):969-80. doi: 10.1017/s1355838202020046. RNA. 2002. PMID: 12212852 Free PMC article. - Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast.
Thakurta AG, Gopal G, Yoon JH, Kozak L, Dhar R. Thakurta AG, et al. EMBO J. 2005 Jul 20;24(14):2512-23. doi: 10.1038/sj.emboj.7600713. Epub 2005 Jun 30. EMBO J. 2005. PMID: 15990877 Free PMC article.
References
- Burge CB, Tuschl TH, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, et al., editors. The RNA world. 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560.
- Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes & Dev. 1997;11:1864–1872. - PubMed
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. New York: Academic Press; 1991.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases