DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals - PubMed (original) (raw)
. 2001 Feb 1;21(3):788-97.
doi: 10.1523/JNEUROSCI.21-03-00788.2001.
C Beard, R Z Chen, G Csankovszki, Y Sun, M Siniaia, D Biniszkiewicz, B Bates, P P Lee, R Kuhn, A Trumpp, C Poon, C B Wilson, R Jaenisch
Affiliations
- PMID: 11157065
- PMCID: PMC6762314
- DOI: 10.1523/JNEUROSCI.21-03-00788.2001
DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals
G Fan et al. J Neurosci. 2001.
Abstract
DNA methyltransferase I (Dnmt1), the maintenance enzyme for DNA cytosine methylation, is expressed at high levels in the CNS during embryogenesis and after birth. Because embryos deficient for Dnmt1 die at gastrulation, the role of Dnmt1 in the development and function of the nervous system could not be studied by using this mutation. We therefore used the cre/loxP system to produce conditional mutants that lack Dnmt1 in neuroblasts of embryonic day 12 embryos or in postmitotic neurons of the postnatal animal. Conditional deletion of the Dnmt1 gene resulted in rapid depletion of Dnmt1 proteins, indicating that the enzyme in postmitotic neurons turns over quickly. Dnmt1 deficiency in postmitotic neurons neither affected levels of global DNA methylation nor influenced cell survival during postnatal life. In contrast, Dnmt1 deficiency in mitotic CNS precursor cells resulted in DNA hypomethylation in daughter cells. Whereas mutant embryos carrying 95% hypomethylated cells in the brain died immediately after birth because of respiratory distress, mosaic animals with 30% hypomethylated CNS cells were viable into adulthood. However, these mutant cells were eliminated quickly from the brain within 3 weeks of postnatal life. Thus, hypomethylated CNS neurons were impaired functionally and were selected against at postnatal stages.
Figures
Fig. 1.
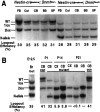
Conditional deletion of the Dnmt1_gene in postmitotic cerebellar neurons. A, Schematic drawing of the cre/loxP-mediated Dnmt1 gene deletion. In the Dnmt1 2lox allele exons 4 and 5 were flanked by the 34 bp loxP sequence. In the presence of cre-recombinase the two loxP sites recombined, resulting in deletion of the exons and creation of the_Dnmt11lox null allele.B, P6 _Dnmt12lox/2lox_cerebellar dissociates were infected with recombinant adenovirus carrying the cre transgene at the time of plating and then were cultured for 1 d (1d) to 14 d (14d). DNAs were extracted from the cultures and digested with Spe_I for Southern blot analysis of the efficiency of the Dnmt1 gene deletion in cerebellar cultures over time. The ratio of recombined null allele (1lox) over the sum of functional_Dnmt12lox (2lox) and 1lox alleles indicates the efficiency of gene deletion.C, Western blot analysis of Dnmt1 proteins in control and mutant neurons. Top, Levels of Dnmt1 protein were detected with Dnmt1 antibodies in cultured cerebellar neurons with or without adeno-cre infection. Bottom, A duplicate gel was stained with Coomassie blue staining to show the equal loading of the protein extracts. D, Southern blot analysis of DNA methylation in cerebellar cultures. DNAs were digested with the methyl-sensitive enzyme _Hpa_II, separated on agarose gel, transferred to the membrane, and hybridized with a centromeric minor satellite repeat probe. Small-molecular-weight fragments in Dnmt1 mutant embryonic stem cells indicate extensive demethylation of their DNA.
Fig. 2.
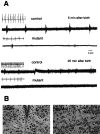
Survival of cerebellar neurons in the absence of Dnmt1. P6 Dnmt1 2lox/2lox cerebellar dissociates were infected with adeno-cre at the beginning and were cultured for 2 weeks. These neuron-enriched cultures were double-stained with neuron-specific β-tubulin III (TuJ1) antibodies (red) to visualize extensive neurites and with DAPI (blue) to show the healthy neuronal nuclei. Scale bar, 45 μm.
Fig. 3.
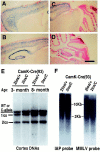
Survival of cortical neurons in the absence of Dnmt1 in vivo. A–D, X-gal staining of brain sections from a 3-week-old mouse carrying the CamK-cre transgene and a lacZ reporter gene under the control of the β-actin promoter (Akagi et al., 1997). The cells positive for β-gal enzymes (blue cells) represent those neurons having undergone cre-mediated gene recombination events. Scale bar, 675 μm.E, Southern blot analysis of cortex DNAs from conditional CamK-cre;Dnmt 2lox/+(2lox/+) heterozygous and_CamK-cre;Dnmt12lox/C_ mutant animals (2lox/C). Note that the wild-type (WT) and null C-alleles were detected at the same size in this blot. The genotypes of wild-type and C-alleles were ascertained by the absence and presence of the neomycin gene in the Dnmt1 locus. F, Methylation analysis in cortex DNAs. IAP, Intra-cisternal A particle retrovirus; MMLV, Moloney murine leukemia virus.
Fig. 4.
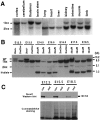
Deletion of the Dnmt1 gene in the brain by the paternally inherited nestin-cre transgene.A, Southern blot analysis of the efficiency of the nestin-cre-mediated Dnmt1 gene deletion in various brain regions and peripheral tissues from E19.5 conditional knock-outs. PhosphorImager analysis of mutant_Dnmt1_ 1lox (1lox) alleles and functional Dnmt12lox(2lox) alleles indicated a recombination efficiency of ∼95% in the brain. Approximately 10–30% recombination was detected in intestine, limb, cranial muscles, and kidney. Only very minor amounts (<5%) of recombination were detected in lung, heart, and liver. B, Dnmt1 gene deletion in E12.5–E18.5 brain tissues. DNA samples were collected from embryos carrying a paternally derived nestin-cre transgene and_Dnmt1_ alleles as indicated (heterozygous control,2lox/+; mutant, 2lox/2lox or_2lox/N_). PhosphorImager analysis showed that ∼95% of the Dnmt12lox allele was recombined into the null Dnmt11lox_allele in the brain from E12.5 to E18.5. C, Western blot analysis of Dnmt1 proteins in control and mutant brain tissues. Brain extracts from E12.5, E15.5, and E18.5 embryos were probed with a specific antibody against the N terminus of the Dnmt1 protein. A duplicate gel was stained with Coomassie blue as a loading control. Note that the E18.5 control sample was from a_Dnmt12lox/N embryo, which showed a reduced level of Dnmt1 proteins as compared with_Dnmt12lox/2lox_ control samples at E12.5 and E15.5. con, Control samples;mut, mutant samples.
Fig. 5.
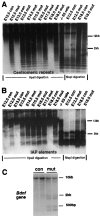
Dnmt1 deficiency results in global genomic hypomethylation. A, B, Southern blot analysis of DNA methylation in the brain. The same DNA samples in Figure1_C_ were used for the methylation assay. DNA was digested with methyl-sensitive enzyme _Hpa_II or methyl-insensitive enzyme Msp_I (-CCGG-) and was hybridized with a centromeric minor repeat probe (A) or an IAP probe (B). The appearance of small-molecular-weight DNA fragments indicates demethylation at the CpG sites of centromeric repeats or IAP retroviruses in the genome.con,Nestin-cre;Dnmt1 +/2lox;mut, Nestin-cre;Dnmt1 2lox/2lox or_2lox/N; −/− ES cells, Demethylated DNA samples from Dnmt1 null embryonic stem cells (Lei et al., 1996). C, Brain DNA from E18.5 embryos was digested with _Sac_I and_Hpa_II enzymes and the probe with a 750 bp BDNF cDNA.
Fig. 6.
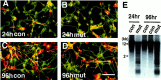
Survival of hypomethylated cortical neurons_in vitro_. A–D, E15.5 cortical cells from control animals (con,Dnmt 2lox/2lox in A and_C_) and_Nestin-cre;Dnmt12lox/2lox_ mutants (mut, in B and D) after 24 or 96 hr in culture. Cultured cells were double-stained with a monoclonal TuJ1 antibody against neuronal-specific β-tubulin III (green) and a polyclonal antibody against nestin intermediate filaments for precursor cells (red). Note that in 24 hr cultures ∼80% of cells were postmitotic neurons. In 96 hr cultures the neurons and glial-like cells (nestin-positive) were ∼50% each, and no difference in these ratios was observed between control and mutant cultures. Scale bar, 60 μm. E, DNA hypomethylation in cultured E15.5 cortical cells. DNAs from cortical cells cultured for 24 or 96 hr were assayed for global demethylation by probing with IAP retro-elements. Demethylated IAP DNA fragments were detected readily in the mutant cultures. con, Control_Dnmt2lox/2lox_ cultures;mut, conditional knock-out cultures.
Fig. 7.
Expression of Xist mRNA in neuronal and glial cells in culture. Xist FISH analysis was performed as described in Materials and Methods. _Xist_RNA expression (red dots in DAPI-stained blue nuclei) was detected in a portion (4–8%) of male mutant cells in 1-d-old E15.5 cortical neuronal cultures (also see Table 1). A few mutant cells also express the Xist transcripts in distributed granules within the cells (third panel), characteristic of cells in the early G1 phase of cell cycle (Clemson et al., 1996).con, Control embryos (Dnmt1 2lox/2lox); mut, conditional mutant embryos. Scale bar, 7 μm.
Fig. 8.
Lack of respiratory drive in the 12th cranial nerve. A, Electrophysiological recordings of descending respiratory discharge from hypoglossal nerve. Control (con, Dnmt1 2lox/2lox) and conditional mutant mice were dissected from the uterus at E18.5 and E19.5 and immediately prepared for 12th nerve recording as described in the Materials and Methods. Only occasional spontaneous gasping was observed in mutant mice. The gasping discharge also could be induced occasionally by tail pinching of mutant mice when an initial recording did not show any spontaneous signals. Similar results were obtained with six control and five mutant mice. Insets for each trace are EKG recordings obtained with subcutaneous electrodes (the EKG in the second control trace was derived from the diaphragmatic EMG recording). B, Normal morphology of hypoglossal motor neurons in the brainstem. E18.5 control (CON) and mutant embryos were fixed with 10% formalin and processed for paraffin histology. Brain sections were stained with cresyl violet. No obvious morphological difference was observed between control and mutant hypoglossal motor nuclei. Scale bar, 37.5 μm.
Fig. 9.
Postnatal loss of Dnmt1-deficient brain cells in mutant mice with maternal inheritance of the nestin-cre transgene.A, Southern blot analysis of the Dnmt1_gene deletion in DNA from different brain regions of mutant mice after maternal inheritance of the nestin-cre transgene. At the newborn stage ∼30–35% of Dnmt1 gene deletion was detected in both mutant and heterozygous brains. FB, Forebrain;Col, colliculus; CB, cerebellum;BS, brainstem; SP, spinal cord.B, Dnmt1-deficient brain cells are eliminated during postnatal development. Deletion of the Dnmt1 gene was maximal by E12.5 (Br, whole brain) and remained constant in the brain throughout the late stage of embryogenesis and at the postnatal day 1 (P1; also see A). However, only a very small number of cells (4–6%) carrying the_Dnmt1 deletion was detected in the cortex (CX) and cerebellum (CB) in 2-week-old (P14) mutant mice. By P21, Dnmt1-deficient cells were not detectable by Southern blot analysis in the cortex, cerebellum, or other regions of the brain (data not shown).mutant, Mutant mice with the_Nestin-cre;Dnmt1_ 2lox/N genotype.con, Control samples from_Nestin-cre;Dnmt1+/2lox_ mice, which showed constant levels of the Dnmt1 deletion at P1 and P21.
Similar articles
- Dnmt1-dependent DNA methylation is essential for photoreceptor terminal differentiation and retinal neuron survival.
Rhee KD, Yu J, Zhao CY, Fan G, Yang XJ. Rhee KD, et al. Cell Death Dis. 2012 Nov 22;3(11):e427. doi: 10.1038/cddis.2012.165. Cell Death Dis. 2012. PMID: 23171847 Free PMC article. - DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation.
Hutnick LK, Golshani P, Namihira M, Xue Z, Matynia A, Yang XW, Silva AJ, Schweizer FE, Fan G. Hutnick LK, et al. Hum Mol Genet. 2009 Aug 1;18(15):2875-88. doi: 10.1093/hmg/ddp222. Epub 2009 May 10. Hum Mol Genet. 2009. PMID: 19433415 Free PMC article. - DNA Methyltransferase 1 Is Indispensable for Development of the Hippocampal Dentate Gyrus.
Noguchi H, Murao N, Kimura A, Matsuda T, Namihira M, Nakashima K. Noguchi H, et al. J Neurosci. 2016 Jun 1;36(22):6050-68. doi: 10.1523/JNEUROSCI.0512-16.2016. J Neurosci. 2016. PMID: 27251626 Free PMC article. - Effects of cerebral ischemia in mice lacking DNA methyltransferase 1 in post-mitotic neurons.
Endres M, Fan G, Meisel A, Dirnagl U, Jaenisch R. Endres M, et al. Neuroreport. 2001 Dec 4;12(17):3763-6. doi: 10.1097/00001756-200112040-00032. Neuroreport. 2001. PMID: 11726790 - DNMT1: catalytic and non-catalytic roles in different biological processes.
Mohan KN. Mohan KN. Epigenomics. 2022 May;14(10):629-643. doi: 10.2217/epi-2022-0035. Epub 2022 Apr 12. Epigenomics. 2022. PMID: 35410490 Review.
Cited by
- DNA Methylation-Dependent Dysregulation of GABAergic Interneuron Functionality in Neuropsychiatric Diseases.
Linde J, Zimmer-Bensch G. Linde J, et al. Front Neurosci. 2020 Sep 16;14:586133. doi: 10.3389/fnins.2020.586133. eCollection 2020. Front Neurosci. 2020. PMID: 33041771 Free PMC article. Review. - Methylation as a key regulator of Tau aggregation and neuronal health in Alzheimer's disease.
Balmik AA, Chinnathambi S. Balmik AA, et al. Cell Commun Signal. 2021 May 7;19(1):51. doi: 10.1186/s12964-021-00732-z. Cell Commun Signal. 2021. PMID: 33962636 Free PMC article. Review. - Proteins in DNA methylation and their role in neural stem cell proliferation and differentiation.
Sun J, Yang J, Miao X, Loh HH, Pei D, Zheng H. Sun J, et al. Cell Regen. 2021 Mar 2;10(1):7. doi: 10.1186/s13619-020-00070-4. Cell Regen. 2021. PMID: 33649938 Free PMC article. Review. - RNA methyltransferase NSun2 deficiency promotes neurodegeneration through epitranscriptomic regulation of tau phosphorylation.
Kim YA, Siddiqui T, Blaze J, Cosacak MI, Winters T, Kumar A, Tein E, Sproul AA, Teich AF, Bartolini F, Akbarian S, Kizil C, Hargus G, Santa-Maria I. Kim YA, et al. Acta Neuropathol. 2023 Jan;145(1):29-48. doi: 10.1007/s00401-022-02511-7. Epub 2022 Nov 10. Acta Neuropathol. 2023. PMID: 36357715 Free PMC article. - Administration of branched-chain amino acids alters epigenetic regulatory enzymes in an animal model of Maple Syrup Urine Disease.
Streck EL, Bussular FP, Wessler LB, Duarte MB, Rezende VL, Rodrigues MS, Torres CA, Lemos IS, Candiotto G, Gava FF, de Oliveira J, Valvassori SS. Streck EL, et al. Metab Brain Dis. 2021 Feb;36(2):247-254. doi: 10.1007/s11011-020-00631-1. Epub 2020 Oct 24. Metab Brain Dis. 2021. PMID: 33098071
References
- Amir RE, Van Den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG binding protein 2. Nat Genet. 1999;23:185–188. - PubMed
- Armstrong DD, Dunn JK, Schultz RJ, Herbert DA, Glaze DG, Motil KJ. Organ growth in Rett syndrome: a postmortem examination analysis. Pediatr Neurol. 1999;20:125–129. - PubMed
- Austin CP, Cepko CL. Cellular migration patterns in the developing mouse cerebral cortex. Development. 1990;110:713–732. - PubMed
- Bates B, Rios M, Trumpp A, Chen C, Fan G, Bishop JM, Jaenisch R. Neurotrophin-3 is required for proper cerebellar development. Nat Neurosci. 1999;2:115–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL52925/HL/NHLBI NIH HHS/United States
- HD18184/HD/NICHD NIH HHS/United States
- R35 CA44339/CA/NCI NIH HHS/United States
- HL60064/HL/NHLBI NIH HHS/United States
- R01 HD018184/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases