Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells - PubMed (original) (raw)
Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells
P Kannouche et al. Genes Dev. 2001.
Abstract
DNA polymerase eta carries out translesion synthesis past UV photoproducts and is deficient in xeroderma pigmentosum (XP) variants. We report that poleta is mostly localized uniformly in the nucleus but is associated with replication foci during S phase. Following treatment of cells with UV irradiation or carcinogens, it accumulates at replication foci stalled at DNA damage. The C-terminal third of poleta is not required for polymerase activity. However, the C-terminal 70 aa are needed for nuclear localization and a further 50 aa for relocalization into foci. Poleta truncations lacking these domains fail to correct the defects in XP-variant cells. Furthermore, we have identified mutations in two XP variant patients that leave the polymerase motifs intact but cause loss of the localization domains.
Figures
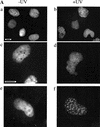
Figure 1
Polη relocalizes into intranuclear foci after UVC-irradiation. (A) MRC5 cells were transfected with plasmids encoding either eGFP-polη (a–d) or untagged polη (e–f). Twenty hours after transfection, cells were irradiated with 7 J/m2 (b,d,f). Eight hours later, the distribution of eGFP-polη was examined after paraformaldehyde fixation (a,b) or in living cells (c,d). For cells transfected with pCDNA-polη, polη distribution was revealed with anti-polη pAb and FITC-conjugated secondary antibody (e,f). Bar = 10 μm. (B) Twenty hours after transfection of MRC5 cells with plasmid encoding eGFP-polη, cells were irradiated with 7 J/m2 UV and then incubated for 8 h. Distribution of eGFP-polη was examined either after paraformaldehyde fixation followed by Triton X-100 permeabilization (a) or with permeabilization carried out before fixation (b). The diffuse staining of eGFP-polη disappeared when cells were preextracted in 1% Triton X-100 before fixation, whereas eGFP-polη foci remain.
Figure 1
Polη relocalizes into intranuclear foci after UVC-irradiation. (A) MRC5 cells were transfected with plasmids encoding either eGFP-polη (a–d) or untagged polη (e–f). Twenty hours after transfection, cells were irradiated with 7 J/m2 (b,d,f). Eight hours later, the distribution of eGFP-polη was examined after paraformaldehyde fixation (a,b) or in living cells (c,d). For cells transfected with pCDNA-polη, polη distribution was revealed with anti-polη pAb and FITC-conjugated secondary antibody (e,f). Bar = 10 μm. (B) Twenty hours after transfection of MRC5 cells with plasmid encoding eGFP-polη, cells were irradiated with 7 J/m2 UV and then incubated for 8 h. Distribution of eGFP-polη was examined either after paraformaldehyde fixation followed by Triton X-100 permeabilization (a) or with permeabilization carried out before fixation (b). The diffuse staining of eGFP-polη disappeared when cells were preextracted in 1% Triton X-100 before fixation, whereas eGFP-polη foci remain.
Figure 2
The relocalization of polη is specific for certain types of DNA lesions. MRC5 (A,B,E–G) or XP12RO (XP-A) (C,D) cells were transfected with eGFP-polη and incubated for 20 h. Cells were then UV irradiated with 7 J/m2 (A,C) and incubated for different times or (B,D) irradiated with different UV doses and incubated for 8 h. The proportion of eGFPpolη-containing cells in which the polη was localized in foci was determined. (E–G) Transfected cells were treated with 5-Gy γ rays, 0.01% MMS, or 100 μM N-AAAF and then incubated for the indicated times before analysis for foci. Top of panel E shows an example of lack of foci after γ irradiation. All panels show representatives of three experiments.
Figure 3
Foci result from relocalization of existing Polη. (A) MRC5 cells were transfected with peGFP-polη and UV irradiated, and individual living cells were examined by time-lapse fluorescence microscopy. Accumulation of polη into foci at 120–200 min is associated with a decrease in intensity of uniform staining throughout the nucleus. (B) XP12RO cells transfected with peGFP-polη were UV irradiated (7 J/m2) and incubated for different times with or without 20 μg/mL cycloheximide. The proportion of cells with foci was determined. (Black bars) Without cycloheximide; (gray bars) with cycloheximide.
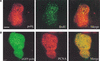
Figure 4
Polη is associated with the replication machinery and partially colocalizes with Rad51. (A) MRC5 cells transfected with pcDNA-polη and pulse labeled with BrdU for 15 min before fixation were double stained with anti-BrdU mAb (green staining) and anti-polη pAb (red staining). (B) Cells transfected with eGFP-polη were fixed and stained with anti-PCNA mAb and TRITC-conjugated secondary antibody. The staining pattern of PCNA (red staining) and the autofluorescent signal of GFP (green staining) in the same cell are shown. Colocalization between BrdU and polη (A) or PCNA and eGFP-polη (B) is indicated by a yellow pattern (right panels). Bar = 5 μm. (C,D) Partial colocalization between polη and Rad51 proteins after UVC irradiation. Twenty hours after transfection with peGFP-polη, cells were UV irradiated with 15 J/m2. Eight hours later, they were fixed and stained with anti-Rad51 pAb and TRITC-conjugated secondary antibody (red staining). The distribution of polη was detected by autofluorescence of the GFP (green staining). In ∼20% of transfected cells, complete colocalization between Rad51 and eGFP-polη foci was observed as shown by yellow staining where red and green signals overlap (C, right panel). In most of the cells, there was a significant but partial colocalization of Rad51 and eGFP-polη foci, as shown by arrows in D. Bar = 5 μm.
Figure 4
Polη is associated with the replication machinery and partially colocalizes with Rad51. (A) MRC5 cells transfected with pcDNA-polη and pulse labeled with BrdU for 15 min before fixation were double stained with anti-BrdU mAb (green staining) and anti-polη pAb (red staining). (B) Cells transfected with eGFP-polη were fixed and stained with anti-PCNA mAb and TRITC-conjugated secondary antibody. The staining pattern of PCNA (red staining) and the autofluorescent signal of GFP (green staining) in the same cell are shown. Colocalization between BrdU and polη (A) or PCNA and eGFP-polη (B) is indicated by a yellow pattern (right panels). Bar = 5 μm. (C,D) Partial colocalization between polη and Rad51 proteins after UVC irradiation. Twenty hours after transfection with peGFP-polη, cells were UV irradiated with 15 J/m2. Eight hours later, they were fixed and stained with anti-Rad51 pAb and TRITC-conjugated secondary antibody (red staining). The distribution of polη was detected by autofluorescence of the GFP (green staining). In ∼20% of transfected cells, complete colocalization between Rad51 and eGFP-polη foci was observed as shown by yellow staining where red and green signals overlap (C, right panel). In most of the cells, there was a significant but partial colocalization of Rad51 and eGFP-polη foci, as shown by arrows in D. Bar = 5 μm.
Figure 5
Recruitment of polη to DNA lesions after UV irradiation. Twenty hours after transfection of MRC5 cells with pCDNA-polη, cells were locally irradiated using a filter. Twelve hours later, polη was detected using anti-polη pAb and FITC-conjugated secondary antibody (green staining), and UV-induced damage in the same cell was revealed using anti-TMD2 mAb and TRITC-conjugated secondary antibody (red staining). Most of the polη foci colocalize with UV lesions, as shown in the right panel (yellow staining). Bar = 5 μm.
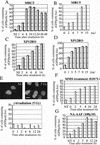
Figure 6
The C-terminal domain of polη is required for the nuclear localization and relocalization after UVC irradiation. (A) Predicted proteins encoded by mutated polη gene in two XP-V cell lines, XP37BR and XP1AB. (B) GFP-tagged deletion mutants are depicted schematically, and the name of each truncated protein is indicated on the left. The conserved domain (aa 1–352) is indicated as well as two putative nuclear localization signals deduced from the amino acid sequence of polη at positions 251–256 and 682–698. (C) Western blots of MRC5 cells transfected with plasmids encoding truncated proteins listed above the panel. Numbers on the left indicate apparent molecular weights in kilodaltons. (D) MRC5 cells were transiently transfected with plasmids expressing GFP-polη642n (a,b), GFP-polη362c (c,d), GFP-polη70c (e,f), and GFP-polη119c (g,h). Twenty hours later, cells were irradiated with 7 J/m2 (b,d,f,h). After 8 h incubation, the localization of the truncated polη proteins was determined by the autofluorescent signal of GFP. Bar = 10 μm.
Figure 6
The C-terminal domain of polη is required for the nuclear localization and relocalization after UVC irradiation. (A) Predicted proteins encoded by mutated polη gene in two XP-V cell lines, XP37BR and XP1AB. (B) GFP-tagged deletion mutants are depicted schematically, and the name of each truncated protein is indicated on the left. The conserved domain (aa 1–352) is indicated as well as two putative nuclear localization signals deduced from the amino acid sequence of polη at positions 251–256 and 682–698. (C) Western blots of MRC5 cells transfected with plasmids encoding truncated proteins listed above the panel. Numbers on the left indicate apparent molecular weights in kilodaltons. (D) MRC5 cells were transiently transfected with plasmids expressing GFP-polη642n (a,b), GFP-polη362c (c,d), GFP-polη70c (e,f), and GFP-polη119c (g,h). Twenty hours later, cells were irradiated with 7 J/m2 (b,d,f,h). After 8 h incubation, the localization of the truncated polη proteins was determined by the autofluorescent signal of GFP. Bar = 10 μm.
Figure 7
Summary of polη domains involved in its cellular localization and bypass activity. (*) Expected results from original truncated protein isolated by Masutani et al. (1999a) with bypass activity comparable to the wild-type protein. (C) Cytoplasmic; (N) nuclear; (C/N) both cytoplasmic and nuclear. (nd) Not determined.
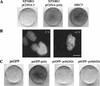
Figure 8
eGFP-polη is functional in vivo and complements XP-variant cells. (A) XP30RO cells were transfected with pCDNA.3 (left) or pCDNA3-polη and incubated for 30 h. The plates were then exposed to 7 J/m2 UV irradiation and incubated in the presence of 75 μg/mL caffeine. After 4 d, the cells were stained with methylene blue. Right panel: MRC5 cells mock transfected and then treated in the same way. (B) XP30RO cells were transfected with peGFP-polη, and stable clones were isolated. UV irradiation results in relocalization of the eGFP-polη like in normal cells. (C) XP30RO cells were transfected with plasmids containing the indicated inserts and then treated as in A. Only the full-length plasmid corrects the UV + caffeine sensitivity.
Figure 9
Alignment of the C-terminal sequences of polη and Rad30 orthologs. (A) The C-terminal 100 aa of human polη (hpolη) are aligned with those of orthologs from mouse (mpolη), Drosophila (dmpolη), Schizosaccharomyces pombe (Eso1), and Saccharomyces cerevisiae (scrad30). Identical aa are indicated in black, conserved aa in gray. The NLS and putative C2H2 zinc finger motifs are indicated by bars above the alignment. (B) Putative PCNA binding motifs in polη are aligned with those from other PCNA-binding proteins (Montecucco et al. 1998).
Similar articles
- Molecular analysis of mutations in DNA polymerase eta in xeroderma pigmentosum-variant patients.
Broughton BC, Cordonnier A, Kleijer WJ, Jaspers NG, Fawcett H, Raams A, Garritsen VH, Stary A, Avril MF, Boudsocq F, Masutani C, Hanaoka F, Fuchs RP, Sarasin A, Lehmann AR. Broughton BC, et al. Proc Natl Acad Sci U S A. 2002 Jan 22;99(2):815-20. doi: 10.1073/pnas.022473899. Epub 2002 Jan 2. Proc Natl Acad Sci U S A. 2002. PMID: 11773631 Free PMC article. - Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells.
Kannouche P, Fernández de Henestrosa AR, Coull B, Vidal AE, Gray C, Zicha D, Woodgate R, Lehmann AR. Kannouche P, et al. EMBO J. 2002 Nov 15;21(22):6246-56. doi: 10.1093/emboj/cdf618. EMBO J. 2002. PMID: 12426396 Free PMC article. Corrected and republished. - Requirement for functional DNA polymerase eta in genome-wide repair of UV-induced DNA damage during S phase.
Auclair Y, Rouget R, Belisle JM, Costantino S, Drobetsky EA. Auclair Y, et al. DNA Repair (Amst). 2010 Jul 1;9(7):754-64. doi: 10.1016/j.dnarep.2010.03.013. Epub 2010 Apr 24. DNA Repair (Amst). 2010. PMID: 20457011 - Xeroderma pigmentosum variant and error-prone DNA polymerases.
Kannouche P, Stary A. Kannouche P, et al. Biochimie. 2003 Nov;85(11):1123-32. doi: 10.1016/j.biochi.2003.10.009. Biochimie. 2003. PMID: 14726018 Review. - Molecular mechanisms of UV-induced mutations as revealed by the study of DNA polymerase eta in human cells.
Stary A, Sarasin A. Stary A, et al. Res Microbiol. 2002 Sep;153(7):441-5. doi: 10.1016/s0923-2508(02)01343-8. Res Microbiol. 2002. PMID: 12405351 Review.
Cited by
- Caffeine and human DNA metabolism: the magic and the mystery.
Kaufmann WK, Heffernan TP, Beaulieu LM, Doherty S, Frank AR, Zhou Y, Bryant MF, Zhou T, Luche DD, Nikolaishvili-Feinberg N, Simpson DA, Cordeiro-Stone M. Kaufmann WK, et al. Mutat Res. 2003 Nov 27;532(1-2):85-102. doi: 10.1016/j.mrfmmm.2003.08.012. Mutat Res. 2003. PMID: 14643431 Free PMC article. - Y-family DNA polymerases in mammalian cells.
Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Guo C, et al. Cell Mol Life Sci. 2009 Jul;66(14):2363-81. doi: 10.1007/s00018-009-0024-4. Epub 2009 Apr 15. Cell Mol Life Sci. 2009. PMID: 19367366 Free PMC article. Review. - Proteomic Profiling Reveals a Specific Role for Translesion DNA Polymerase η in the Alternative Lengthening of Telomeres.
Garcia-Exposito L, Bournique E, Bergoglio V, Bose A, Barroso-Gonzalez J, Zhang S, Roncaioli JL, Lee M, Wallace CT, Watkins SC, Opresko PL, Hoffmann JS, O'Sullivan RJ. Garcia-Exposito L, et al. Cell Rep. 2016 Nov 8;17(7):1858-1871. doi: 10.1016/j.celrep.2016.10.048. Cell Rep. 2016. PMID: 27829156 Free PMC article. - Mutations in human DNA polymerase eta motif II alter bypass of DNA lesions.
Glick E, Vigna KL, Loeb LA. Glick E, et al. EMBO J. 2001 Dec 17;20(24):7303-12. doi: 10.1093/emboj/20.24.7303. EMBO J. 2001. PMID: 11743006 Free PMC article. - The unusual UBZ domain of Saccharomyces cerevisiae polymerase η.
Woodruff RV, Bomar MG, D'Souza S, Zhou P, Walker GC. Woodruff RV, et al. DNA Repair (Amst). 2010 Nov 10;9(11):1130-41. doi: 10.1016/j.dnarep.2010.08.001. Epub 2010 Sep 15. DNA Repair (Amst). 2010. PMID: 20837403 Free PMC article.
References
- Arlett CF, Harcourt SA. Survey of radiosensitivity in a variety of human cell strains. Cancer Res. 1980;40:926–932. - PubMed
- Arlett CF, Harcourt SA, Broughton BC. The influence of caffeine on cell survival in excision-proficient and excision-deficient xeroderma pigmentosum and normal human cell strains following ultraviolet light irradiation. Mutation Res. 1975;33:341–346. - PubMed
- Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: A potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes & Dev. 1994;8:811–820. - PubMed
- Berneburg M, Lehmann AR. Advances in Genetics. Vol. 43. San Diego, CA: Academic Press; 2001. Xeroderma pigmentosum and related disorders: Defects in DNA repair and transcription; pp. 71–102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous