Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains - PubMed (original) (raw)
Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains
C Hennequin et al. J Clin Microbiol. 2001 Feb.
Abstract
Since Saccharomyces cerevisiae appears to be an emerging pathogen, there is a need for a valuable molecular marker able to distinguish among strains. In this work, we investigated the potential value of microsatellite length polymorphism with a panel of 91 isolates, including 41 clinical isolates, 14 laboratory strains, and 28 strains with industrial relevance. Testing seven polymorphic regions (five trinucleotide repeats and two dinucleotide repeats) in a subgroup of 58 unrelated strains identified a total of 69 alleles (6 to 13 per locus) giving 52 different patterns with a discriminatory power of 99.03%. We found a cluster of clinical isolates sharing their genotype with a bakery strain, suggesting a digestive colonization following ingestion of this strain with diet. With the exception of this cluster of isolates and isolates collected from the same patient or from patients treated with Saccharomyces boulardii, all clinical isolates gave different and unique patterns. The genotypes are stable, and the method is reproducible. The possibility to make the method portable is of great interest for further studies using this technique. This work shows the possibility to readily identify S. boulardii (a strain increasingly isolated from invasive infections) using a unique and specific microsatellite allele.
Figures
FIG. 1
Denaturing polyacrylamide gel electrophoresis of products of PCR amplification of the YKL172w microsatellite (microsatellite 1). S. cerevisiae strains are described in Table 1. Note that CLIB 1260 (lane 3) corresponds to an S. pastorianus strain. *, clinical isolates that were collected from patients taking S. boulardii therapy.
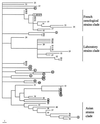
FIG. 2
Strict consensus of the 1,479 most-parsimonious phylogenetic trees. The phylogram is constructed using the midpoint rooting method. Tree length, 208; consistency index, 0.332; retention index, 0.7. Strain numbering refers to Table 1. Clinical isolates from patients not taking S. boulardii are circled. ∗ (boxed), S. boulardii reference strain (1), isolates from S. boulardii commercial preparations (90 and 91), and isolates from patients treated with S. boulardii (69 to 89).
Similar articles
- Molecular tools for differentiating probiotic and clinical strains of Saccharomyces cerevisiae.
Posteraro B, Sanguinetti M, Romano L, Torelli R, Novarese L, Fadda G. Posteraro B, et al. Int J Food Microbiol. 2005 Sep 15;103(3):295-304. doi: 10.1016/j.ijfoodmicro.2004.12.031. Int J Food Microbiol. 2005. PMID: 16099314 - Species and genotypic diversities and similarities of pathogenic yeasts colonizing women.
Xu J, Boyd CM, Livingston E, Meyer W, Madden JF, Mitchell TG. Xu J, et al. J Clin Microbiol. 1999 Dec;37(12):3835-43. doi: 10.1128/JCM.37.12.3835-3843.1999. J Clin Microbiol. 1999. PMID: 10565893 Free PMC article. - Selection of hypervariable microsatellite loci for the characterization of Saccharomyces cerevisiae strains.
Legras JL, Ruh O, Merdinoglu D, Karst F. Legras JL, et al. Int J Food Microbiol. 2005 Jun 25;102(1):73-83. doi: 10.1016/j.ijfoodmicro.2004.12.007. Int J Food Microbiol. 2005. PMID: 15925004 - A database of microsatellite genotypes for Saccharomyces cerevisiae.
Richards KD, Goddard MR, Gardner RC. Richards KD, et al. Antonie Van Leeuwenhoek. 2009 Oct;96(3):355-9. doi: 10.1007/s10482-009-9346-3. Epub 2009 Apr 26. Antonie Van Leeuwenhoek. 2009. PMID: 19396625 - Molecular characterization of clinical Saccharomyces cerevisiae isolates and their association with non-clinical strains.
de Llanos R, Querol A, Planes AM, Fernández-Espinar MT. de Llanos R, et al. Syst Appl Microbiol. 2004 Aug;27(4):427-35. doi: 10.1078/0723202041438473. Syst Appl Microbiol. 2004. PMID: 15368848
Cited by
- Structure and features of the complete chloroplast genome of Melastoma dodecandrum.
Zheng X, Ren C, Huang S, Li J, Zhao Y. Zheng X, et al. Physiol Mol Biol Plants. 2019 Jul;25(4):1043-1054. doi: 10.1007/s12298-019-00651-x. Epub 2019 Mar 12. Physiol Mol Biol Plants. 2019. PMID: 31404219 Free PMC article. - Identification of Geotrichum candidum at the species and strain level: proposal for a standardized protocol.
Gente S, Sohier D, Coton E, Duhamel C, Gueguen M. Gente S, et al. J Ind Microbiol Biotechnol. 2006 Dec;33(12):1019-31. doi: 10.1007/s10295-006-0130-3. Epub 2006 Jul 20. J Ind Microbiol Biotechnol. 2006. PMID: 16855820 - Comparative genomic study on the complete plastomes of four officinal Ardisia species in China.
Xie C, An W, Liu S, Huang Y, Yang Z, Lin J, Zheng X. Xie C, et al. Sci Rep. 2021 Nov 15;11(1):22239. doi: 10.1038/s41598-021-01561-3. Sci Rep. 2021. PMID: 34782652 Free PMC article. - Molecular genotyping of Candida parapsilosis group I clinical isolates by analysis of polymorphic microsatellite markers.
Lasker BA, Butler G, Lott TJ. Lasker BA, et al. J Clin Microbiol. 2006 Mar;44(3):750-9. doi: 10.1128/JCM.44.3.750-759.2006. J Clin Microbiol. 2006. PMID: 16517850 Free PMC article. - Comparison of restriction fragment length polymorphism, microsatellite length polymorphism, and random amplification of polymorphic DNA analyses for fingerprinting Aspergillus fumigatus isolates.
Bart-Delabesse E, Sarfati J, Debeaupuis JP, van Leeuwen W, van Belkum A, Bretagne S, Latge JP. Bart-Delabesse E, et al. J Clin Microbiol. 2001 Jul;39(7):2683-6. doi: 10.1128/JCM.39.7.2683-2686.2001. J Clin Microbiol. 2001. PMID: 11427596 Free PMC article.
References
- Aucott J N, Fayen J, Grossnicklas H, Morrissey A, Lederman M M, Salata R A. Invasive infection with Saccharomyces cerevisiae: report of three cases and review. Rev Infect Dis. 1990;12:406–411. - PubMed
- Boekhout T, van Belkum A, Leenders A C, Verbrugh H A, Mukamurangwa P, Swinne D, Scheffers W A. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int J Syst Bacteriol. 1997;47:432–442. - PubMed
- Clemons K V, McCusker J H, Davis R W, Stevens D A. Comparative pathogenesis of clinical and nonclinical isolates of Saccharomyces cerevisiae. J Infect Dis. 1994;169:859–867. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases