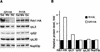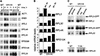Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome biosynthesis - PubMed (original) (raw)
Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome biosynthesis
T L Iouk et al. Mol Cell Biol. 2001 Feb.
Abstract
Ribosome biogenesis is regulated by environmental cues that coordinately modulate the synthesis of ribosomal components and their assembly into functional subunits. We have identified an essential yeast WD-repeat-containing protein, termed Rrb1p, that has a role in both the assembly of the 60S ribosomal subunits and the transcriptional regulation of ribosomal protein (RP) genes. Rrb1p is located in the nucleus and is concentrated in the nucleolus. Its presence is required to maintain normal cellular levels of 60S subunits, 80S ribosomes, and polyribosomes. The function of Rrb1p in ribosome biogenesis appears to be linked to its association with the ribosomal protein rpL3. Immunoprecipitation of Rrb1p from nuclear extracts revealed that it physically interacts with rpL3. Moreover, the overproduction of Rrb1p led to increases in cellular levels of free rpL3 that accumulated in the nucleus together with Rrb1p. The concentration of these proteins within the nucleus was dependent on ongoing protein translation. We also showed that overexpression of RRB1 led to an increase in the expression of RPL3 while all other examined RP genes were unaffected. In contrast, depletion of RRB1 caused an increase in the expression of all RP genes examined except RPL3. These results suggest that Rrb1p regulates RPL3 expression and uncouples it from the coordinated expression of other RP genes.
Figures
FIG. 1
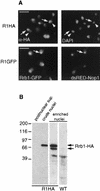
Rrb1p is located in the nucleus and concentrated in the nucleolus. (A) R1HA cells were fixed, permeabilized, and probed with MAb 12CA5. Binding was detected with rhodamine-conjugated, goat anti-mouse antibodies. Nuclear DNA was visualized by DAPI staining. The corresponding positions of MAb 12CA5 (arrows) and DAPI (arrowheads) binding in several nuclei are shown. Below these are shown images of the strain R1GFP-N expressing an integrated RRB1-GFP construct and plasmid-borne DsRED-NOP1 fusion. Rrb1-GFP and nucleolar DsRED-Nop1 were detected using fluorescein isothiocyanate and rhodamine filters, respectively, and the arrow points to the same region of the nucleus in both images. Bar, 5 μm. (B) Rrb1p cofractionates with nuclei. R1HA cells were used to produce a postnuclear supernatant, a crude nuclear pellet, and an enriched nuclear fraction. Rrb1-HA in these fractions was detected by Western analysis using MAb 12CA5. Wild-type (WT) W303 nuclei lacking the Rrb1-HA were also probed with MAb 12CA5. The positions of molecular mass markers are shown in kilodaltons.
FIG. 2
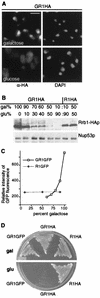
GAL1 regulation of RRB1 expression. (A) GR1HA (GAL1::RRB1-HA) cells were grown in medium containing a galactose: glucose mixture (60:40) and then shifted to either 2% galactose- or 2% glucose-containing medium for 6 h. Rrb1-HA was detected by immunofluorescence as described for Fig. 1, using MAb 12CA5. Nuclear DNA was visualized by DAPI staining. Bar, 5 μm. (B) The levels of Rrb1-HA were examined in GR1HA (GAL1::RRB1-HA) and R1HA (RRB1 controlled by its endogenous promoter) cells grown with various carbon sources. Cells were grown in a 2% carbon source composed of the proportion of galactose (gal) and glucose (glu) indicated. Western blotting was performed on total cell lysates derived from equal amounts of cells, using MAb 12CA5 to detect Rrb1-HA or an antibody directed against the nuclear pore complex protein Nup53p (31). (C) Cultures of GR1GFP and R1GFP (RRB1-GFP controlled by the RRB1 promoter) were grown to early logarithmic phase and the mean fluorescent intensity (y axis) of 104 cells was determined by fluorescence-activated cell sorter analysis and plotted versus the percentage of galactose in the carbon source mixture. (D) GR1GFP, GR1HA, and R1HA cells were grown overnight in media containing a 60:40 mixture of galactose and glucose and then plated onto YP plates containing either galactose or glucose. Plates were incubated for 2 days at 30°C.
FIG. 3
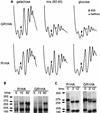
The effect of depletion of Rrb1p on ribosome biosynthesis. (A) R1HA and GR1HA (GAL1::RRB1-HA) cells were grown in medium containing a galactose:glucose (60:40) mixture and then shifted to medium containing either 2% galactose or 2% glucose for 4 h. Ribosomal subunits, ribosomes, and polyribosomes derived from these cultures were then separated on 7-to-47% sucrose gradients and detected by their UV absorbance at 254 nm. The position of the 60S ribosomal subunit is indicated by an arrow. Half-mers are highlighted with an asterisk. (B and C) Pulse-chase analysis of rRNA. R1HA and GR1HA cells were grown in glucose-containing medium for 4 h, pulse-labeled with [5, 6-3H]uridine (B) or with [_methyl_-3H]methionine (C) for 3 min, and chased with medium lacking the labeling reagent. Following the indicated chase time, total RNA was isolated, resolved on agarose gels, and detected by autoradiography. The positions of the various rRNA species are indicated according to their sedimentation coefficients.
FIG. 4
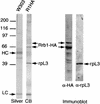
The ribosomal protein rpL3 is associated with Rrb1p. Nuclei isolated from R1HA and W303 (lacking an HA tag) strains were extracted with 1% Triton X-100 and 240 mM NaCl. Cleared supernatants were incubated with MAb 12CA5 conjugated to protein G-Sepharose beads to immunoprecipitate Rrb1-HA. The proteins eluted from these beads were then resolved using SDS-PAGE and detected with either silver or Coomassie blue (CB) staining. Immunoprecipitates derived from the R1HA nuclei were also analyzed by Western blotting using MAb 12CA5 (α-HA) and the MAb TCM1 directed against the ribosomal protein rpL3 (α-rpL3). Of the three major bands visible in the eluate, the 70- and 66-kDa species bound MAb 12CA5, and the 40-kDa species bound the α-rpL3 antibody. The positions of light chain (LC) and heavy chain (HC) species derived from MAb 12CA5 are indicated. The positions of molecular mass markers are shown in kilodaltons.
FIG. 5
Overproduction of Rrb1p leads to an increase in cellular levels of rpL3. (A) GR1HA (GAL1::RRB1-HA) cells were grown in galactose:glucose (70:30)-containing media (Mix) and then shifted to media containing either galactose (Gal) or glucose (Glc) for 6 h. Total cell lysates were isolated and equal amounts of protein were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with MAb 12CA5 (Rrb1-HA), MAb TCM1 (rpL3), anti-rpL30 (rpL30 and rpS2), and anti-Nup53p (Nup53p). The ribosomal protein rpS2 was detected as a result of its cross-reactivity with anti-rpL30 antibody (60). Switching to galactose resulted in increases in the cellular levels of Rrb1p and rpL3 but not rpL30, rpS2, or Nup53p. (B) GR1HA (GAL1::RRB1-HA) and R1HA (RRB1-HA under the control of its endogenous promoter) cells were grown in galactose-containing medium, and equal amounts of total cell lysates were analyzed by Western blotting using antibodies directed against the indicated proteins. Signals were quantified as described in Materials and Methods. The signals detected in the R1HA strain were assigned a value of one. The fold increases in the GR1HA strain of the individual proteins relative to their amounts in the R1HA strain were plotted.
FIG. 6
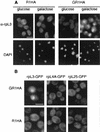
rpL3 accumulates in the nuclei of cells overexpressing RRB1. (A) GR1HA (GAL1:RRB1-HA) and R1HA cells were grown in galactose:glucose (70:30)-containing medium and shifted to either galactose or glucose for 6 h. Cells were then fixed, permeabilized, and probed with the MAb TCM1 (α-rpL3). Nuclear DNA was visualized by DAPI staining. (B) GR1HA and R1HA cells containing a plasmid-borne copy of the gene fusion RPL3-GFP, RPL4A-GFP, or RPL25-GFP were grown in medium containing a galactose:glucose (70:30) mixture and then shifted to galactose-containing medium for 6 h. The GFP fusions were visualized directly by fluorescence microscopy. The position of the nuclei within these cells was determined by Nomarski optics (data not shown). Bar, 5 μm.
FIG. 7
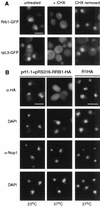
Inhibition of protein synthesis causes a reversible relocalization of Rrb1p from the nucleus to the cytoplasm. (A) GR1GFP (GAL1:RRB1-GFP) cells (Rrb1-GFP) and GR1HA (GAL1:RRB1-HA) cells expressing RPL3-GFP (rpL3-GFP) were grown in galactose-containing medium, and the GFP fusions were visualized by fluorescence microscopy (untreated). These cells were then treated with cycloheximide (100 μg/μl) for 1.5 h and reexamined by fluorescence microscopy (+CHX). Following treatment with cycloheximide, cells were washed, resuspended in fresh medium, and allowed to recover for 30 min at 30°C (CHX removed). (B) R1HA cells and the thermosensitive prt1-1 strain containing pRS316-RRB1-HA were grown at 23°C and then shifted to 37°C for 20 min. Cells harvested from both temperatures were fixed, permeabilized, and probed with either MAb 12CA5 (α-HA) or an MAb that specifically binds the nucleolar protein Nop1p (α-Nop1). Nuclear DNA was visualized by DAPI staining. Bar, 5 μm.
FIG. 8
Rrb1p modulates RP mRNA levels. (A) GR1HA (GAL1:RRB1-HA) and R1HA (WT) cells were grown in galactose:glucose (70:30)-containing medium and then shifted to either galactose-or glucose-containing media. At the indicated time points, total RNA was isolated. Equal amounts of RNA from each of these samples were separated on a 1.2% formaldehyde-agarose gel, transferred to a nylon membrane, and hybridized with the indicated 32P-labeled cDNA probe. Note that the RRB1 blot was exposed for a time period similar to that for the ribosomal protein mRNA blots and thus it is only visible here in the overexpressing samples. (B) Northern blots were performed as for panel A, and signals were quantitated using a phosphorimager. mRNA levels were normalized to ACT1 mRNA and wild-type levels of RP mRNAs were assigned a value of one. (C) Northern analysis was performed on RNA isolated from GR1HA cells expressing RPL25-GFP or RPL3-GFP driven by an exogenous promoter (see Materials and Methods). Both mRNAs were detected using a 32P-labeled RPL25 or RPL3 cDNA probe. (D) The effects of a GAL10::NOP1 conditional allele on the levels of RPL25 mRNA were examined in strain Y159. Y159 (GAL10::NOP1) and W303 (WT) cells were grown in galactose:glucose (70:30)-containing medium and then shifted to either galactose- or glucose-containing medium for 12 h. Northern analysis was performed on total RNA using a 32P-labeled RPL25 cDNA probe. The lane order is the same as that shown in panel C.
Similar articles
- Factors affecting nuclear export of the 60S ribosomal subunit in vivo.
Stage-Zimmermann T, Schmidt U, Silver PA. Stage-Zimmermann T, et al. Mol Biol Cell. 2000 Nov;11(11):3777-89. doi: 10.1091/mbc.11.11.3777. Mol Biol Cell. 2000. PMID: 11071906 Free PMC article. - Inactivation of the RRB1-Pescadillo pathway involved in ribosome biogenesis induces chromosomal instability.
Killian A, Le Meur N, Sesboüé R, Bourguignon J, Bougeard G, Gautherot J, Bastard C, Frébourg T, Flaman JM. Killian A, et al. Oncogene. 2004 Nov 11;23(53):8597-602. doi: 10.1038/sj.onc.1207845. Oncogene. 2004. PMID: 15467761 - Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p.
Zanchin NI, Goldfarb DS. Zanchin NI, et al. Mol Cell Biol. 1999 Feb;19(2):1518-25. doi: 10.1128/MCB.19.2.1518. Mol Cell Biol. 1999. PMID: 9891085 Free PMC article. - Ribosome synthesis in Saccharomyces cerevisiae.
Venema J, Tollervey D. Venema J, et al. Annu Rev Genet. 1999;33:261-311. doi: 10.1146/annurev.genet.33.1.261. Annu Rev Genet. 1999. PMID: 10690410 Review. - Nuclear export and cytoplasmic maturation of ribosomal subunits.
Zemp I, Kutay U. Zemp I, et al. FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
Cited by
- A RanGTP-independent mechanism allows ribosomal protein nuclear import for ribosome assembly.
Schütz S, Fischer U, Altvater M, Nerurkar P, Peña C, Gerber M, Chang Y, Caesar S, Schubert OT, Schlenstedt G, Panse VG. Schütz S, et al. Elife. 2014 Aug 21;3:e03473. doi: 10.7554/eLife.03473. Elife. 2014. PMID: 25144938 Free PMC article. - Histidine methylation of yeast ribosomal protein Rpl3p is required for proper 60S subunit assembly.
Al-Hadid Q, Roy K, Munroe W, Dzialo MC, Chanfreau GF, Clarke SG. Al-Hadid Q, et al. Mol Cell Biol. 2014 Aug;34(15):2903-16. doi: 10.1128/MCB.01634-13. Epub 2014 May 27. Mol Cell Biol. 2014. PMID: 24865971 Free PMC article. - Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits.
West M, Hedges JB, Chen A, Johnson AW. West M, et al. Mol Cell Biol. 2005 May;25(9):3802-13. doi: 10.1128/MCB.25.9.3802-3813.2005. Mol Cell Biol. 2005. PMID: 15831484 Free PMC article. - The p21-activated protein kinase inhibitor Skb15 and its budding yeast homologue are 60S ribosome assembly factors.
Saveanu C, Rousselle JC, Lenormand P, Namane A, Jacquier A, Fromont-Racine M. Saveanu C, et al. Mol Cell Biol. 2007 Apr;27(8):2897-909. doi: 10.1128/MCB.00064-07. Epub 2007 Feb 16. Mol Cell Biol. 2007. PMID: 17308036 Free PMC article. - A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity.
Cloutier P, Lavallée-Adam M, Faubert D, Blanchette M, Coulombe B. Cloutier P, et al. PLoS Genet. 2013;9(1):e1003210. doi: 10.1371/journal.pgen.1003210. Epub 2013 Jan 17. PLoS Genet. 2013. PMID: 23349634 Free PMC article.
References
- Aitchison J D, Rout M P, Marelli M, Blobel G, Wozniak R W. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. - PMC - PubMed
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1992.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases