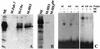The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity - PubMed (original) (raw)
The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity
R K Mishra et al. Mol Cell Biol. 2001 Feb.
Abstract
In the work reported here we have undertaken a functional dissection of a Polycomb response element (PRE) from the iab-7 cis-regulatory domain of the Drosophila melanogaster bithorax complex (BX-C). Previous studies mapped the iab-7 PRE to an 860-bp fragment located just distal to the Fab-7 boundary. Located within this fragment is an approximately 230-bp chromatin-specific nuclease-hypersensitive region called HS3. We have shown that HS3 is capable of functioning as a Polycomb-dependent silencer in vivo, inducing pairing-dependent silencing of a mini-white reporter. The HS3 sequence contains consensus binding sites for the GAGA factor, a protein implicated in the formation of nucleosome-free regions of chromatin, and Pleiohomeotic (Pho), a Polycomb group protein that is related to the mammalian transcription factor YY1. We show that GAGA and Pho interact with these sequences in vitro and that the consensus binding sites for the two proteins are critical for the silencing activity of the iab-7 PRE in vivo.
Figures
FIG. 1
Defining the minimal PRE and mutations in GAGA binding sites. The Fab-7 boundary and iab-7 PRE region of the BX-C are shown at the top. These two elements lie adjacent to one another, and both map between the PS11 regulatory domain, iab-6, on the left and the PS12 regulatory domain, iab-7, on the right. The Bluetail transposon, BLT, which is subject to the control of iab-7 but not iab-6, lies between hypersensitive sites HS2 and HS3 (16). The 860-, 410-, and 260-bp fragments containing HS3 are shown. The number of pairing-sensitive lines out of the total number of transgenic lines of each construct is shown to the right. Stars, GAGA consensus binding sites; small boxes, Pho consensus binding sites. The bottom construct depicts the 260-bp fragment containing mutations in both of the GAGA consensus binding sites.
FIG. 2
GAGA and Pho proteins bind to recognition sequences in HS3. (A) Binding of GAGA protein to the iab-7 PRE. In this experiment nuclear extract was incubated with matrix containing different DNA sequences. The first was a control sequence from a region of Fab-7 that does not contain any consensus GAGA protein recognition sequences. The second was a fragment from the Ubx promoter, which contains four consensus GAGA recognition sequences. The third was the 260-bp iab-7 PRE fragment spanning HS3 (see text and Fig. 1). It contains two consensus GAGA recognition sequences. Proteins bound to each matrix were eluted with increasing salt concentrations. Shown is the 0.4 M KCl wash. The protein samples eluted from each matrix were separated by SDS–10% PAGE and transferred on a PVDF membrane for Western analysis with an anti-GAGA antibody. Similar quantities of nuclear extract and affinity matrix were used in order to compare the relative abundances of protein in each fraction. While multiple isoforms of GAGA (plus presumptive breakdown products) are detected by the antibody in nuclear extracts, only a subset of these bind to the matrix. For the HS3 matrix, the major species is the lowest band, which corresponds to the 70-kDa isoform, while larger isoforms are generally present in lower yield. Note that little or no GAGA protein appears to be bound to the Fab-7 matrix, which does not contain consensus GAGA binding sites. M-Fab-7, matrix containing Fab-7 DNA; M-Ubx, matrix containing Ubx promoter DNA; M-HS3, matrix containing HS3 DNA. (B) Binding of GAGA protein to the iab-7 PRE depends on the two consensus GAGA binding sites. We compared the binding of the GAGA factor to an affinity matrix containing the wild-type HS3 sequence and a matrix containing the HS3 sequence with mutations in the two consensus GAGA binding sites (see text and Fig. 1). The 70-kDa isoform binds to the matrix containing the wild-type HS3 sequence (M-HS3) but does not bind to the matrix containing the HS3 sequence with GAGA binding site mutations (M-HS3ga). (C) Labeled double-stranded oligonucleotide probes (see Materials and Methods). wt, wild type; m9, mutated at position 9 (G to T); mc, mutated at core positions 2, 3, and 4 (CAT to ACG). Two to five micrograms of protein extract (NE, nuclear extract; BE, bacterial extract expressing Pho) was incubated with the labeled DNA and cold carrier (poly[dI-dC] and tRNA) in binding buffer at room temperature for 30 min. The mixture was then analyzed on a 4% acrylamide-bisacrylamide (80:1) gel in 0.5× TBE containing 2.5% glycerol. Specificity was confirmed by competition experiments in which a 100-fold molar excess of either wild-type or mutant oligonucleotides was included in the binding mixture. The core mutant was found to be a much weaker competitor than the wild-type sequence. To a lesser extent this was also true of the +9 mutant (data not shown).
FIG. 3
Mutations in Pho consensus binding sites. Black bar, 860-bp iab-7 PRE _Apa_I-_Xba_I fragment; large box, hypersensitive site 3; stars and small boxes, GAGA and Pho consensus binding sites, respectively. The 860-bp iab-7 PRE fragment was inserted into an enhancer-less mini-white transgene. Portions of the sequence for each of the fragments tested are shown. The number of pairing-sensitive lines out of the total number of transgenic lines is shown for each construct. (A) Wild-type sequence; (B) 15-bp deletion that removes the proximal Pho consensus binding site; (C) 60-bp deletion that removes both Pho consensus binding sites and the intervening sequence; (D) point mutations generated in the Pho core consensus sequence; (E) point mutations generated in both of the +9 conserved residues. Boldface, conserved sequences within the Pho consensus binding site; italics, GAGA consensus sites. For the point mutations (D and E), the mutated sequence is shown in boldface above the residues that were mutated.
FIG. 4
Pho is required for the silencing activity of the iab-7 PRE. Shown is the eye color phenotype of one of the 860-bp iab-7 PRE mini-white transgenic lines, P24, in different genetic backgrounds. As a hemizygote in a wild-type background, this insert produces a light orange eye color phenotype. When the transgene is homozygous, mini-white expression is repressed, and the eye color of the P24/P24 animals is almost white. The pho gene is required for the iab-7 PRE-dependent silencing of mini-white. This can be seen by comparing the eye color of P24/P24; pho1/phocv animals with that of wild-type P24/P24 animals.
Similar articles
- A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex.
Hagstrom K, Muller M, Schedl P. Hagstrom K, et al. Genetics. 1997 Aug;146(4):1365-80. doi: 10.1093/genetics/146.4.1365. Genetics. 1997. PMID: 9258680 Free PMC article. - The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1.
Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. Brown JL, et al. Mol Cell. 1998 Jun;1(7):1057-64. doi: 10.1016/s1097-2765(00)80106-9. Mol Cell. 1998. PMID: 9651589 - In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element.
Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. Mihaly J, et al. Development. 1997 May;124(9):1809-20. doi: 10.1242/dev.124.9.1809. Development. 1997. PMID: 9165128 - Heritable chromatin states induced by the Polycomb and trithorax group genes.
Paro R, Strutt H, Cavalli G. Paro R, et al. Novartis Found Symp. 1998;214:51-61; discussion 61-6, 104-13. doi: 10.1002/9780470515501.ch4. Novartis Found Symp. 1998. PMID: 9601011 Review. - Chromatin. Ga-ga over GAGA factor.
Granok H, Leibovitch BA, Shaffer CD, Elgin SC. Granok H, et al. Curr Biol. 1995 Mar 1;5(3):238-41. doi: 10.1016/s0960-9822(95)00048-0. Curr Biol. 1995. PMID: 7780729 Review.
Cited by
- Boundary bypass activity in the abdominal-B region of the Drosophila bithorax complex is position dependent and regulated.
Kyrchanova O, Ibragimov A, Postika N, Georgiev P, Schedl P. Kyrchanova O, et al. Open Biol. 2023 Aug;13(8):230035. doi: 10.1098/rsob.230035. Epub 2023 Aug 16. Open Biol. 2023. PMID: 37582404 Free PMC article. - Transcriptional Readthrough Interrupts Boundary Function in Drosophila.
Kyrchanova O, Sokolov V, Tikhonov M, Manukyan G, Schedl P, Georgiev P. Kyrchanova O, et al. Int J Mol Sci. 2023 Jul 12;24(14):11368. doi: 10.3390/ijms241411368. Int J Mol Sci. 2023. PMID: 37511131 Free PMC article. - Boundary Bypass Activity in the Abdominal-B Region of the Drosophila Bithorax Complex is Position Dependent and Regulated.
Kyrchanova O, Ibragimov A, Postika N, Georgiev P, Schedl P. Kyrchanova O, et al. bioRxiv [Preprint]. 2023 Jun 7:2023.06.06.543971. doi: 10.1101/2023.06.06.543971. bioRxiv. 2023. PMID: 37333165 Free PMC article. Updated. Preprint. - Distinct roles for canonical and variant histone H3 lysine-36 in Polycomb silencing.
Salzler HR, Vandadi V, McMichael BD, Brown JC, Boerma SA, Leatham-Jensen MP, Adams KM, Meers MP, Simon JM, Duronio RJ, McKay DJ, Matera AG. Salzler HR, et al. Sci Adv. 2023 Mar;9(9):eadf2451. doi: 10.1126/sciadv.adf2451. Epub 2023 Mar 1. Sci Adv. 2023. PMID: 36857457 Free PMC article. - Transcriptional read through interrupts boundary function in Drosophila.
Kyrchanova O, Sokolov V, Tikhonov M, Schedl P, Georgiev P. Kyrchanova O, et al. bioRxiv [Preprint]. 2023 Feb 16:2023.02.16.528790. doi: 10.1101/2023.02.16.528790. bioRxiv. 2023. PMID: 36824960 Free PMC article. Updated. Preprint.
References
- Barges S, Mihaly J, Galloni M, Hagstrom K, Müller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. - PubMed
- Bender W, Akam M, Karch F, Beachy P A, Peifer M, Spierer P, Lewis E B, Hogness D S. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science. 1983;221:23–29. - PubMed
- Bhat K M, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1113–1124. - PubMed
- Biggin M D, Tjian R. Transcription factors that activate the Ubx promoter in developmentally staged extracts. Cell. 1988;53:699–711. - PubMed
- Boulet A, Lloyd A, Sakonju S. Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Development. 1991;111:393–405. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases