Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation - PubMed (original) (raw)
Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation
K Mattsson et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Several recent findings have indicated that the promyelocytic leukemia gene product (PML) oncogenic domains (PODs) are involved in proteasome-mediated degradation of ubiquitinated proteins. We wanted to examine the intracellular distribution of PML protein in the presence of a proteasome inhibitor. We used high-resolution microscopy to study the distribution of PML protein and other POD-associated proteins along with the proteasomes themselves under normal conditions and in cells treated with the proteasome inhibitor, MG132. Inhibition of the proteasomes in MCF-7, HeLa, and IB-4 cell lines resulted in a radical redistribution of the POD-associated proteins PML, Sp100, and SUMO-1. After 6-10 h of MG132 treatment, PML, Sp100, and SUMO-1 were no longer detectable in the PODs and accumulated mainly in the nucleolus. Moreover, MG132 treatment changed the cellular distribution of the proteasomes. Interestingly, this included the accumulation in euchromatin areas of the nucleus and within the nucleoli. Several non-POD-associated proteins did not change their cellular distribution under the same conditions. The accumulation of POD-associated proteins and proteasomes in the nucleoli of MG132-treated cells indicates that these proteins may target the nucleoli under normal conditions and that the nucleolus may have a function in the regulation of proteasomal protein degradation.
Figures
Figure 1
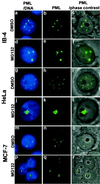
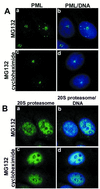
Subnuclear distribution of PML after DMSO or MG132 treatment of MCF-7, HeLa, or LCL IB-4 cell lines. Combination of DNA (blue) and PML (green) is shown in the left panels (a, d,g, j, m,p), middle panels show PML alone (b,e, h, k, n,q), combination of phase-contrast and immunofluorescence of PML is shown in right panels (c, f,i, l, o,r). IB-4 cells cultured in the presence of DMSO (a–c) with PML distributed throughout the nucleoplasm excluding the nucleoli. Cells cultured with 5 μM MG132 for 6 h (d–f) show staining of PML inside the nucleoli (d, f) in addition to some nuclear dots. HeLa cells cultured in the presence of DMSO (g–i) or with 5 μM MG132 for 15 h (j–l). PML accumulation upon MG132 treatment is shown in one of three nucleoli (l). MCF-7 cells cultured in the presence of DMSO (m–o) or with 5 μM MG132 for 15 h (p–r). PML accumulates in the nucleoli of MG132-treated cells (r).
Figure 2
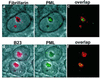
Nucleolar localization of PML in MG132-treated MCF-7 cells. High magnification image of double staining for fibrillarin and PML or for B23 and PML. (a) Phase-contrast field combined with fibrillarin staining (red). (b) Phase-contrast and nucleolar PML (green) staining representing the same field as shown in_a_. (c) Overlap between fibrillarin and PML staining. (d) Phase-contrast field combined with nucleolar B23 staining (red). Panels e [phase-contrast and nucleolar PML staining (green)] and f overlap between B23 and PML.
Figure 3
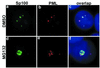
Sp100 changes subcellular distribution and disassociates from PML upon MG132 treatment of MCF-7 cells. High magnification of Sp100 (green) and PML (red) double-staining of DMSO (a–c) or MG132-treated MCF-7 cells (d–f). (c and f) Overlap of Sp100 and PML. In DMSO-treated cells, Sp100 colocalized to a large extent with PML (c). After MG132 treatment, Sp100 completely changed its localization (d and f); it accumulated in the nucleoli along with PML but without any colocalization (f). DNA staining in blue.
Figure 4
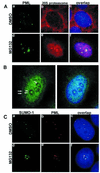
(A) Double staining of PML (green) and proteasomes (red) in DMSO- or MG132-treated MCF-7 cells. In DMSO-treated cells, proteasomes were homogeneously distributed throughout the nucleus excluding nucleoli (b). Double staining showed no obvious colocalization between the two proteins (c). MG132 treatment changed the nuclear distribution of both PML and proteasomes; they both accumulated in the nucleoli as shown in panel (d–f). (B) Subnuclear localization of proteasomes in MG132-treated MCF-7 cells. Proteasomes (green) accumulate in the euchromatin areas and nucleoli and avoid peripheral (solid arrowheads) and perinucleolar (concave arrowheads) heterochromatin. (C) a–c represent double staining for SUMO-1 (green) and PML (red) in DMSO-treated MCF-7 cells. Overlap image shows complete colocalization of the two proteins (c). Upon MG132 treatment, SUMO-1 and PML accumulated in the nucleoli (d and e) without colocalization. DNA staining in blue.
Figure 5
Three-dimensional stereoscopic reconstitution of PML (red) and SUMO-1 (green) double-stained cell (MCF-7) treated with MG132 showing that although both proteins accumulated in the nucleolus, they do not colocalize with each other any longer. The mathematically deblurred image was generated from a series of 11 optical sections, 0.3 μm apart. DNA staining in blue.
Figure 6
Effect of cycloheximide on the nucleolar accumulation of PML and proteasomes. (A) MCF-7 cells that were treated with MG132 alone (a and b) or with MG132 plus cycloheximide (c and d) over night did not change the nucleolar accumulation of PML (green). (B) The nucleolar accumulation of proteasomes (green) was not altered by the inhibition of protein synthesis in MCF-7 cells (a–d). DNA staining in blue.
Similar articles
- Interferon gamma regulates accumulation of the proteasome activator PA28 and immunoproteasomes at nuclear PML bodies.
Fabunmi RP, Wigley WC, Thomas PJ, DeMartino GN. Fabunmi RP, et al. J Cell Sci. 2001 Jan;114(Pt 1):29-36. doi: 10.1242/jcs.114.1.29. J Cell Sci. 2001. PMID: 11112687 - Proteasome-dependent processing of nuclear proteins is correlated with their subnuclear localization.
Dino Rockel T, von Mikecz A. Dino Rockel T, et al. J Struct Biol. 2002 Oct-Dec;140(1-3):189-99. doi: 10.1016/s1047-8477(02)00527-0. J Struct Biol. 2002. PMID: 12490167 - Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression.
Xu Y, Ahn JH, Cheng M, apRhys CM, Chiou CJ, Zong J, Matunis MJ, Hayward GS. Xu Y, et al. J Virol. 2001 Nov;75(22):10683-95. doi: 10.1128/JVI.75.22.10683-10695.2001. J Virol. 2001. PMID: 11602710 Free PMC article. - PML and COP1--two proteins with much in common.
Reyes JC. Reyes JC. Trends Biochem Sci. 2001 Jan;26(1):18-20. doi: 10.1016/s0968-0004(00)01732-1. Trends Biochem Sci. 2001. PMID: 11165511 Review. - Intracellular localization of proteasomes.
Wójcik C, DeMartino GN. Wójcik C, et al. Int J Biochem Cell Biol. 2003 May;35(5):579-89. doi: 10.1016/s1357-2725(02)00380-1. Int J Biochem Cell Biol. 2003. PMID: 12672451 Review.
Cited by
- Characterization and functional analysis of chicken promyelocytic leukemia protein.
Wang S, Li J, Miao T, Li T, Wan Z, Xie Q, Shao H, Qin A, Ye J. Wang S, et al. Poult Sci. 2024 Dec;103(12):104272. doi: 10.1016/j.psj.2024.104272. Epub 2024 Aug 28. Poult Sci. 2024. PMID: 39293264 Free PMC article. - Dictyostelium discoideum has a highly Q/N-rich proteome and shows an unusual resilience to protein aggregation.
Malinovska L, Palm S, Gibson K, Verbavatz JM, Alberti S. Malinovska L, et al. Proc Natl Acad Sci U S A. 2015 May 19;112(20):E2620-9. doi: 10.1073/pnas.1504459112. Epub 2015 May 4. Proc Natl Acad Sci U S A. 2015. PMID: 25941378 Free PMC article. - Topological stress triggers persistent DNA lesions in ribosomal DNA with ensuing formation of PML-nucleolar compartment.
Urbancokova A, Hornofova T, Novak J, Salajkova SA, Stemberkova Hubackova S, Uvizl A, Buchtova T, Mistrik M, McStay B, Hodny Z, Bartek J, Vasicova P. Urbancokova A, et al. Elife. 2024 Oct 10;12:RP91304. doi: 10.7554/eLife.91304. Elife. 2024. PMID: 39388244 Free PMC article. - Non-canonical role of wild-type SEC23B in the cellular stress response pathway.
Yehia L, Liu D, Fu S, Iyer P, Eng C. Yehia L, et al. Cell Death Dis. 2021 Mar 22;12(4):304. doi: 10.1038/s41419-021-03589-9. Cell Death Dis. 2021. PMID: 33753724 Free PMC article. - Post-translational modifications of PML: consequences and implications.
Cheng X, Kao HY. Cheng X, et al. Front Oncol. 2013 Jan 4;2:210. doi: 10.3389/fonc.2012.00210. eCollection 2012. Front Oncol. 2013. PMID: 23316480 Free PMC article.
References
- Seeler J S, Dejean A. Curr Opin Genet Dev. 1999;9:362–367. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources