Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus - PubMed (original) (raw)
Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus
M Kihara et al. J Bacteriol. 2001 Mar.
Abstract
The MS ring of the flagellar basal body of Salmonella is an integral membrane structure consisting of about 26 subunits of a 61-kDa protein, FliF. Out of many nonflagellate fliF mutants tested, three gave rise to intergenic suppressors in flagellar region II. The pseudorevertants swarmed, though poorly; this partial recovery of motile function was shown to be due to partial recovery of export function and flagellar assembly. The three parental mutants were all found to carry the same mutation, a six-base deletion corresponding to loss of Ala-174 and Ser-175 in the predicted periplasmic domain of the FliF protein. The 19 intergenic suppressors identified all lay in flhA, and they consisted of 10 independent examples at the nucleotide level or 9 at the amino acid level. Since two of the nine corresponded to different substitutions at the same amino acid position, only eight positions in the FlhA protein have given rise to suppressors. Thus, FliF-FlhA intergenic suppression is a fairly rare event. FlhA is a component of the flagellar protein export apparatus, with an integral membrane domain encompassing the N-terminal half of the sequence and a cytoplasmic C-terminal domain. All of the suppressing mutations lay within the integral membrane domain. These mutations, when placed in a wild-type fliF background, had no mutant phenotype. In the fliF mutant background, mutant FlhA was dominant, yielding a pseudorevertant phenotype. Wild-type FlhA did not exert significant negative dominance in the pseudorevertant background, indicating that it does not compete effectively with mutant FlhA for interaction with mutant FliF. Mutant FliF was partially dominant over wild-type FliF in both the wild-type and second-site FlhA backgrounds. Membrane fractionation experiments indicated that the fliF mutation, though preventing export, was mild enough to permit assembly of the MS ring itself, and also assembly of the cytoplasmic C ring onto the MS ring. The data from this study provide genetic support for a model in which at least the FlhA component of the export apparatus physically interacts with the MS ring within which it is housed.
Figures
FIG. 1
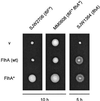
Swarming abilities of SJW1103 (wild type [wt]), SJW2706 (parental fliF mutant, _fliF_∗), MM0608 (pseudorevertant, _fliF_∗ _flhA_∗), and MM1608 (_flhA_∗ single-site mutant derived from MM0608). The plates were incubated for the times indicated; note the much longer times used for the _fliF_∗ mutant and the pseudorevertant than for the wild type and the _flhA_∗ strain.
FIG. 2
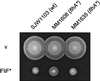
Swarm test of the complementation and dominance properties of wild-type (wt) FlhA (middle row) and suppressor mutant FlhA (FlhA∗) from pseudorevertant MM0608. pTrc99A-based plasmids producing these proteins were used to transform the strains indicated at the top. SJW2706 is the parental mutant (fliF*), MM0608 is a pseudorevertant (fliF* flhA*), and SJW1364 is a flhA null mutant. v, pTrc99A vector. Transformed cells were incubated on tryptone semisolid agar plates for the times indicated.
FIG. 3
Swarm test of the dominance properties of mutant FliF (FliF∗) from SJW2706. A pTrc99A-based plasmid, pMMiF2706, producing this protein was used to transform the strains indicated at the top. SJW1103 is wild type (wt); MM1608 and MM1635 are second-site mutants (flhA*). v, pTrc99A vector. Transformed cells were incubated on tryptone semisolid agar plates for 5 h at 30°C.
FIG. 4
Distribution of the MS-ring protein FliF and the C-ring motor/switch proteins FliG and FliN between the high-speed pellet, or membrane, fraction (m) and the supernatant fraction (s) of cells of wild-type SJW1103 (wild type [wt]), a fliF null mutant SJW1684 (fliF), the first-site fliF mutant SJW2706 (_fliF_∗), and a flhA null mutant SJW1364 (flhA). The positions of molecular mass markers (in kilodaltons) are shown on the left.
FIG. 5
Export of the hook-capping protein FlgD to the periplasm (p) and culture supernatant (s) of cells of wild-type (wt) SJW1103, the first-site fliF mutant SJW2706 (_fliF_∗), the pseudorevertant (_fliF_∗ _flhA_∗), and the flgD null mutant SJW156 (flgD). Samples were subjected to SDS-PAGE and immunoblotted with polyclonal anti-FlgD antibody. Size in kilodaltons is indicated at the left.
FIG. 6
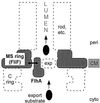
Cartoon interpreting the suppression of a mutational defect in the MS-ring protein FliF by a mutation in one of the membrane components of the flagellar protein export apparatus, FlhA. Other membrane components of the export apparatus are indicated by exp and a dashed oval. The export apparatus, which translocates export substrates from the cytoplasm into the lumen of the nascent flagellar structure (rod, etc.), is believed to be located in a patch of membrane within the pore that exists within the MS ring. Suppression is postulated to be a result of physical interaction (double-headed arrow) between the inner annular surface of the MS ring and the transmembrane region of FlhA or, in one case, near the interface between the transmembrane region and the soluble domain. The C ring is a part of the motor that is mounted onto the MS ring. CM, cytoplasmic membrane; cyto, cytoplasm; peri, periplasm.
FIG. 7
Schematic illustration of the predicted transmembrane organization of FlhA, the integral membrane component of the export apparatus that gave rise to suppression of mutations in the MS-ring protein FliF. Positions of residues at the beginning and end of predicted terminal and loop regions are indicated. Mutations identified in this study are indicated by boldface letters and asterisks. CM, cytoplasmic membrane; cyto, cytoplasm; peri, periplasm.
FIG. 8
Schematic illustration of effects of FliF and FlhA overproduction (as a result of expression from pTrc99A-based plasmids) on swarming in various host backgrounds. (i) Wild-type, untransformed or transformed with vector; (ii) parental FliF∗ mutant, untransformed or transformed with vector; (iii) FliF∗ FlhA∗ pseudorevertant, untransformed or transformed with vector; (iv) second-site FlhA∗ mutant, untransformed or transformed with vector; (v) parental FliF∗ mutant with FliF∗ overexpressed; (vi) FliF∗ FlhA∗ pseudorevertant with FliF∗ overexpressed; (vii) FliF∗ FlhA∗ pseudorevertant with FlhA∗ overexpressed; (viii) wild-type with FliF∗ overexpressed; (ix) second-site FlhA∗ mutant with FliF∗ overexpressed; (x) FliF∗ FlhA∗ pseudorevertant with FlhA overexpressed. F, wild-type FliF; F∗, mutant FliF; A, wild-type FlhA; A∗, mutant FlhA. Chromosomal expression is indicated by the smaller font, while plasmid expression is indicated by the larger font. Swarming ability is indicated qualitatively by the diameter of the black circles, categorized by four levels: wild-type, intermediate, pseudorevertant, and parental. The data are interpreted in terms of a physical interaction between FliF and FlhA subunits, where the strength of the interaction is indicated by the thickness of the connecting bar. Wild-type proteins are indicated by light shading, and mutant proteins are indicated by dark shading. In cases (viii) and (ix), the possibility of a mixed (FliF|FliF∗) multimer is indicated (see text).
Similar articles
- Analysis of the cytoplasmic domains of Salmonella FlhA and interactions with components of the flagellar export machinery.
McMurry JL, Van Arnam JS, Kihara M, Macnab RM. McMurry JL, et al. J Bacteriol. 2004 Nov;186(22):7586-92. doi: 10.1128/JB.186.22.7586-7592.2004. J Bacteriol. 2004. PMID: 15516571 Free PMC article. - Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body.
Morimoto YV, Ito M, Hiraoka KD, Che YS, Bai F, Kami-Ike N, Namba K, Minamino T. Morimoto YV, et al. Mol Microbiol. 2014 Mar;91(6):1214-26. doi: 10.1111/mmi.12529. Epub 2014 Feb 15. Mol Microbiol. 2014. PMID: 24450479 - Type III flagellar protein export and flagellar assembly.
Macnab RM. Macnab RM. Biochim Biophys Acta. 2004 Nov 11;1694(1-3):207-17. doi: 10.1016/j.bbamcr.2004.04.005. Biochim Biophys Acta. 2004. PMID: 15546667 Review. - Suppressor Mutants: History and Today's Applications.
Bautista DE, Carr JF, Mitchell AM. Bautista DE, et al. EcoSal Plus. 2021 Dec 15;9(2):eESP00372020. doi: 10.1128/ecosalplus.ESP-0037-2020. Epub 2021 Dec 15. EcoSal Plus. 2021. PMID: 34910591 Free PMC article. Review.
Cited by
- A Proline-Rich Element in the Type III Secretion Protein FlhB Contributes to Flagellar Biogenesis in the Beta- and Gamma-Proteobacteria.
Hook JC, Blagotinsek V, Pané-Farré J, Mrusek D, Altegoer F, Dornes A, Schwan M, Schier L, Thormann KM, Bange G. Hook JC, et al. Front Microbiol. 2020 Dec 15;11:564161. doi: 10.3389/fmicb.2020.564161. eCollection 2020. Front Microbiol. 2020. PMID: 33384667 Free PMC article. - Structural Conservation and Adaptation of the Bacterial Flagella Motor.
Carroll BL, Liu J. Carroll BL, et al. Biomolecules. 2020 Oct 29;10(11):1492. doi: 10.3390/biom10111492. Biomolecules. 2020. PMID: 33138111 Free PMC article. Review. - Analysis of the cytoplasmic domains of Salmonella FlhA and interactions with components of the flagellar export machinery.
McMurry JL, Van Arnam JS, Kihara M, Macnab RM. McMurry JL, et al. J Bacteriol. 2004 Nov;186(22):7586-92. doi: 10.1128/JB.186.22.7586-7592.2004. J Bacteriol. 2004. PMID: 15516571 Free PMC article. - Interactions between flagellar and type III secretion proteins in Chlamydia pneumoniae.
Stone CB, Bulir DC, Gilchrist JD, Toor RK, Mahony JB. Stone CB, et al. BMC Microbiol. 2010 Jan 22;10:18. doi: 10.1186/1471-2180-10-18. BMC Microbiol. 2010. PMID: 20096108 Free PMC article. - FliH and FliI help FlhA bring strict order to flagellar protein export in Salmonella.
Kinoshita M, Minamino T, Uchihashi T, Namba K. Kinoshita M, et al. Commun Biol. 2024 Mar 26;7(1):366. doi: 10.1038/s42003-024-06081-0. Commun Biol. 2024. PMID: 38531947 Free PMC article.
References
- Fan F, Macnab R M. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. - PubMed
- Fan F, Ohnishi K, Francis N R, Macnab R M. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. - PubMed
- Fraser G M, Bennett J C Q, Hughes C. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol. 1999;32:569–580. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases