The structure of the ultraspiracle ligand-binding domain reveals a nuclear receptor locked in an inactive conformation - PubMed (original) (raw)
The structure of the ultraspiracle ligand-binding domain reveals a nuclear receptor locked in an inactive conformation
G M Clayton et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Ultraspiracle (USP) is the invertebrate homologue of the mammalian retinoid X receptor (RXR). RXR plays a uniquely important role in differentiation, development, and homeostasis through its ability to serve as a heterodimeric partner to many other nuclear receptors. RXR is able to influence the activity of its partner receptors through the action of the ligand 9-cis retinoic acid. In contrast to RXR, USP has no known high-affinity ligand and is thought to be a silent component in the heterodimeric complex with partner receptors such as the ecdysone receptor. Here we report the 2.4-A crystal structure of the USP ligand-binding domain. The structure shows that a conserved sequence motif found in dipteran and lepidopteran USPs, but not in mammalian RXRs, serves to lock USP in an inactive conformation. It also shows that USP has a large hydrophobic cavity, implying that there is almost certainly a natural ligand for USP. This cavity is larger than that seen previously for most other nuclear receptors. Intriguingly, this cavity has partial occupancy by a bound lipid, which is likely to resemble the natural ligand for USP.
Figures
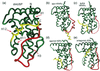
Figure 1
The structure of USP compared with other nuclear receptors. The loop between helices 1 and 3 (red) and helix 12 (yellow) in dmUSP differ from other receptors.
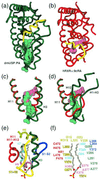
Figure 2
The conserved H1–H3 loop locks USP in an inactive conformation. (a) Interactions between the H1–H3 loop and adjacent parts of the structure. Green dashes indicate hydrogen bonds. (b) Schematic view of the interactions illustrated in_a_. Solid and dashed lines indicate nonpolar van der Waals interactions and hydrogen bonds, respectively. Residues conserved in all dipteran and lepidopteran USPs are colored pink. (c) Sequence alignment of dipteran/lepidopteran USPs and mammalian RXRs. Residues conserved in the holometabolous insect orders (pink) correspond to those residues highlighted in_b_. dm, Drosophila melanogaster; bm,Bombyx mori; ms, Manduca sexta.
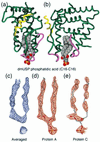
Figure 3
An unexpected ligand for USP. (a and b) Orthogonal views of phosphatidic acid bound to dmUSP. Helix 12 is colored yellow. Magenta loops indicate mobile regions in some LBDs (see text) (c_–_e) 2_F_o −_F_c electron-density maps of the ligand before and after refinement (contoured at 1σ).
Figure 4
An unusual ligand-binding cavity. (a and_b_) dmUSP (green) compared with hRXRα (red). 9cRA (magenta) is smaller than the phosphatidic acid (cyan). Helix 12 is colored yellow. Compared with hRXRα:9cRA, helix 3 is splayed out of the ligand-binding cavity (c), and helix 12 is displaced by the H1–H3 loop (d). (e) Regions of the LBD contributing to the ligand-binding cavity: H1–H3 (yellow), H3 (dark green), H5 (light green), S1–S2 (blue), H6 (magenta), H7 (cyan), and H11 (red). (f) Side chains in contact with ligand; coloring is as in e.
References
- Oro A E, McKeown M, Evans R M. Nature (London) 1990;347:298–301. - PubMed
- Hayward D C, Bastiani M J, Trueman J W, Truman J W, Riddiford L M, Ball E E. Dev Genes Evol. 1999;209:564–571. - PubMed
- Sluder A E, Mathews S W, Hough D, Yin V P, Maina C V. Genome Res. 1999;9:103–120. - PubMed
- Yao T P, Segraves W A, Oro A E, McKeown M, Evans R M. Cell. 1992;71:63–72. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases