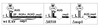A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus - PubMed (original) (raw)
A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus
M A Tijms et al. Proc Natl Acad Sci U S A. 2001.
Abstract
The genome expression of positive-stranded RNA viruses starts with translation rather than transcription. For some viruses, the genome is the only viral mRNA and expression is regulated primarily at the translational level and by limited proteolysis of polyproteins. Other virus groups also generate subgenomic mRNAs later in the reproductive cycle. For nidoviruses, subgenomic mRNA synthesis (transcription) is discontinuous and yields a 5' and 3' coterminal nested set of mRNAs. Nidovirus transcription is not essential for genome replication, which relies on the autoprocessing products of two replicase polyproteins that are translated from the genome. We now show that the N-terminal replicase subunit, nonstructural protein 1 (nsp1), of the nidovirus equine arteritis virus is in fact dispensable for replication but crucial for transcription, thereby coupling replicase expression and subgenomic mRNA synthesis in an unprecedented manner. Nsp1 is composed of two papain-like protease domains and a predicted N-terminal zinc finger, which was implicated in transcription by site-directed mutagenesis. The structural integrity of nsp1 is essential, suggesting that the protease domains form a platform for the zinc finger to operate in transcription.
Figures
Figure 1
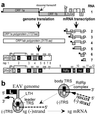
(A) Schematic diagram of EAV genome organization and expression. The nested set of sg mRNAs (RNAs 2 to 7) is shown, with the leader sequence represented as a black box and the ORF(s) expressed from each mRNA depicted in gray. (Lower) The ORF1a and ORF1ab replicase polyproteins and their processing maps are depicted with protease domains and corresponding cleavage sites indicated. Abbreviations for conserved domains: ZF, nonstructural protein- (nsp) 1 zinc finger; α, nsp1 papain-like cysteine protease (PCP) α; β, nsp1 PCPβ; C, nsp2 cysteine protease; H, hydrophobic domain; S, nsp4 serine protease; RdRp, RNA-dependent RNA polymerase; M, predicted metal-binding domain; Hel, helicase; D, conserved nidovirus-specific domain. (B) Model for nidovirus discontinuous minus-strand synthesis, yielding sg minus-strand RNAs that function as a template for sg mRNAs in transcription (9, 10). Discontinuous minus-strand synthesis involves attenuation of the RdRp complex at the body transcription-regulating sequence (TRS), translocation of the nascent minus strand to the leader TRS in the genomic template [exposed by the leader TRS hairpin (LTH)], base pairing between the minus-sense body TRS and plus-sense leader TRS, and reinitiation of RNA synthesis to complete the sg minus strand with the complement of the leader sequence.
Figure 2
Construction of knockout mutant Δnsp1. Depicted is a schematic overview of the important elements in the 5′ end of the EAV genome: the genomic leader sequence (L), LTH, ORF1a and its translation-initiation codon (circle), the nsp1 zinc finger (ZF) and PCP domains (see Fig. 5), and the nsp2 coding region. The replicase ORF is shown as a solid bar.
Figure 3
Nsp1 is an essential factor for transcription but not replication of the EAV genome. (A) Immunoprecipitation analysis of nsp1 (29 kDa) and nsp2 (61 kDa) expression. (B) Northern blot analysis of intracellular RNA (24 h after transfection) with a probe complementary to the 3′ end of all EAV mRNAs (10). The positions of the EAV genome and six sg mRNAs are indicated. The band labeled E in the DITRAC lane represents an extra sg RNA transcribed from a cryptic TRS in the EMCV IRES (see text). (C) RT-PCR analysis of sg-RNA7 synthesis. The generation of minus- (Upper) or plus-stranded (Lower) sg RNA7 in transfected cells was tested by RNA7-specific RT-PCR. One of the primers was located in the RNA7 body and the other in the leader sequence, thereby generating an RNA7-specific PCR product of 540 bp (arrowhead). Mutant EAV030F, which generates about 500-fold reduced levels of sg RNAs (17, 20), was included to confirm the sensitivity of the assay.
Figure 4
Trans-complementation of the function of nsp1 in transcription. (A) Outline of the DITRAC replicon and complementation assay. The expression of nsp1 from the 5′ end of the genome was inactivated (see Fig. 2_A_), and an IRES-nsp1 cassette was inserted in the structural gene region. Complementation of nsp1 function should restore sg-RNA synthesis from the DITRAC 3′ end. (B) Reporter-gene expression from the IRES in control-replicon DICAT, which is fully negative for sg-mRNA synthesis (Fig. 3). (C) Trans-complementation of nsp1 function. Cells were transfected with wild-type EAV030 RNA or with DITRAC, fixed at 24 h after electroporation, and double stained for nsp3 and the nucleocapsid protein to monitor genome replication and transcription, respectively. The nucleocapsid protein is expressed from sg mRNA7 (see_A_).
Figure 5
Identification of a ZF motif in the N-terminal domain of the arterivirus replicase. The sequences of lactate dehydrogenase elevating virus [LDV-C (44) and LDV-P (45)], porcine reproductive and respiratory syndrome virus [PRRSV-LV (46) and PRRSV-VR2332 (47)], and EAV (6) were compared. Alignments of the complete ZF domain and selected conserved regions of PCPα and PCPβ containing the active-site Cys and His residues (bold) are shown. Invariant (*) and conserved (:) positions are highlighted. Conserved His and Cys residues in the nsp1 ZF that are proposed to be involved in zinc binding are shown in reverse shading, and mutated residues (Table 1) are indicated with “#.”
Figure 6
RT-PCR analysis of sg-mRNA7 synthesis by DITRAC derivatives containing either a truncated nsp1 gene or specific point mutations (Table 1) in the ZF, PCPα, or PCPβ domains. See Fig. 3_C_ for RT-PCR details.
Similar articles
- Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease.
Tijms MA, Nedialkova DD, Zevenhoven-Dobbe JC, Gorbalenya AE, Snijder EJ. Tijms MA, et al. J Virol. 2007 Oct;81(19):10496-505. doi: 10.1128/JVI.00683-07. Epub 2007 Jul 11. J Virol. 2007. PMID: 17626105 Free PMC article. - The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis.
van Dinten LC, van Tol H, Gorbalenya AE, Snijder EJ. van Dinten LC, et al. J Virol. 2000 Jun;74(11):5213-23. doi: 10.1128/jvi.74.11.5213-5223.2000. J Virol. 2000. PMID: 10799597 Free PMC article. - Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis.
van Marle G, van Dinten LC, Spaan WJ, Luytjes W, Snijder EJ. van Marle G, et al. J Virol. 1999 Jul;73(7):5274-81. doi: 10.1128/JVI.73.7.5274-5281.1999. J Virol. 1999. PMID: 10364273 Free PMC article. - Nidovirus RNA polymerases: Complex enzymes handling exceptional RNA genomes.
Posthuma CC, Te Velthuis AJW, Snijder EJ. Posthuma CC, et al. Virus Res. 2017 Apr 15;234:58-73. doi: 10.1016/j.virusres.2017.01.023. Epub 2017 Feb 6. Virus Res. 2017. PMID: 28174054 Free PMC article. Review. - Nidovirales: evolving the largest RNA virus genome.
Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Gorbalenya AE, et al. Virus Res. 2006 Apr;117(1):17-37. doi: 10.1016/j.virusres.2006.01.017. Epub 2006 Feb 28. Virus Res. 2006. PMID: 16503362 Free PMC article. Review.
Cited by
- Functional specialization and evolution of leader proteinases in the family Closteroviridae.
Peng CW, Peremyslov VV, Mushegian AR, Dawson WO, Dolja VV. Peng CW, et al. J Virol. 2001 Dec;75(24):12153-60. doi: 10.1128/JVI.75.24.12153-12160.2001. J Virol. 2001. PMID: 11711606 Free PMC article. - RNA transcription analysis and completion of the genome sequence of yellow head nidovirus.
Sittidilokratna N, Dangtip S, Cowley JA, Walker PJ. Sittidilokratna N, et al. Virus Res. 2008 Sep;136(1-2):157-65. doi: 10.1016/j.virusres.2008.05.008. Epub 2008 Jun 11. Virus Res. 2008. PMID: 18582978 Free PMC article. - Sugarcane mosaic virus reduced bacterial diversity and network complexity in the maize root endosphere.
Liu W, Cui X, Wang X, Shen C, Ji L, Zhang M, Wong MH, Zhang J, Shan S. Liu W, et al. mSystems. 2023 Aug 31;8(4):e0019823. doi: 10.1128/msystems.00198-23. Epub 2023 Jun 29. mSystems. 2023. PMID: 37382454 Free PMC article. - The viral innate immune antagonism and an alternative vaccine design for PRRS virus.
Ke H, Yoo D. Ke H, et al. Vet Microbiol. 2017 Sep;209:75-89. doi: 10.1016/j.vetmic.2017.03.014. Epub 2017 Mar 18. Vet Microbiol. 2017. PMID: 28341332 Free PMC article. Review. - Classification, replication, and transcription of Nidovirales.
Liao Y, Wang H, Liao H, Sun Y, Tan L, Song C, Qiu X, Ding C. Liao Y, et al. Front Microbiol. 2024 Jan 24;14:1291761. doi: 10.3389/fmicb.2023.1291761. eCollection 2023. Front Microbiol. 2024. PMID: 38328580 Free PMC article. Review.
References
- Molenkamp R, van Tol H, Rozier B C D, van der Meer Y, Spaan W J M, Snijder E J. J Gen Virol. 2000;81:2491–2496. - PubMed
- Snijder E J, Meulenberg J J M. J Gen Virol. 1998;79:961–979. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials