Familial dysautonomia is caused by mutations of the IKAP gene - PubMed (original) (raw)
Familial dysautonomia is caused by mutations of the IKAP gene
S L Anderson et al. Am J Hum Genet. 2001 Mar.
Abstract
The defective gene DYS, which is responsible for familial dysautonomia (FD) and has been mapped to a 0.5-cM region on chromosome 9q31, has eluded identification. We identified and characterized the RNAs encoded by this region of chromosome 9 in cell lines derived from individuals homozygous for the major FD haplotype, and we observed that the RNA encoding the IkappaB kinase complex-associated protein (IKAP) lacks exon 20 and, as a result of a frameshift, encodes a truncated protein. Sequence analysis reveals a T-->C transition in the donor splice site of intron 20. In individuals bearing a minor FD haplotype, a missense mutation in exon 19 disrupts a consensus serine/threonine kinase phosphorylation site. This mutation results in defective phosphorylation of IKAP. These mutations were observed to be present in a random sample of Ashkenazi Jewish individuals, at approximately the predicted carrier frequency of FD. These findings demonstrate that mutations in the gene encoding IKAP are responsible for FD.
Figures
Figure 1
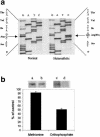
Deletion in IKAP mRNA, resulting in a truncated protein. a, RT-PCR analysis of IKAP RNA. DNase-treated total RNA was prepared, by use of RNAqueous-4PCR kits (Ambion), from lymphoblast cell lines generated from individuals who either lacked the DYS allele (normal) (DY491; generated in our laboratory) or were either heterozygous (carrier) (GM05046A, GM05107, and GM05108A [NIGMS Human Genetic Mutant Cell Repository]) or homozygous (affected) (GM05106 [NIGMS Human Genetic Mutant Cell Repository]) for the major DYS allele. RT-PCR was performed with EZrTth RNA PCR kits (Applied Biosystems) with primers spanning exons 19–21 (5′-GCAGCAATCATGTGTCCCA-3′ and 5′-GATTCTCAGCTTTCTCATGC-3′). The positions of the DNA markers run on the gel are indicated by arrows. b, Alignment of normal and FD IKAP cDNA and amino acid sequences. An alignment of a portion of exons 19–21 of normal IKAP cDNA with that of IKAP cDNA from an FD-affected individual, demonstrating that the exclusion of exon 20 in the RNA transcribed from the FD allele results in a frameshift, causing premature termination of translation and a protein truncated by 619 amino acids; normal IKAP has 1,332 amino acids. c, Western blot analysis of IKAP protein in a non-FD lymphoblast cell line (DY491) and in a lymphoblast cell line established from an FD-affected individual (GM05106). The blot was probed with polyclonal antibody to IKAP (Santa Cruz Biotechnology) prepared, in goat, against the peptide, DPVSREVKNEVSLVAEGF, encoded in exons 2 and 3. The presence of IKAP was detected with an anti-goat antibody conjugated to alkaline phosphatase (Promega). No bands appeared on western blots in which the antibody had been preincubated with the peptide used to generate it (data not shown). A prestained molecular-weight marker was used to determine sizes of products (data not shown).
Figure 2
T→C transition in a donor splice site, resulting in excision of exon 20. a, Sequence analysis of IKAP intron 20 donor splice sites in normal and FD alleles. PCR fragments, which were amplified from DNA derived from normal cells and from cells homozygous for the major FD haplotype and which span the intron 20 donor splice of IKAP, were sequenced. The normal donor splice sequence is GTAAGTG; the FD sequence is GTAAGCG. b, Splicing of the IKAP transcript. Normal splicing of the IKAP transcript results in removal of introns 19 and 20 and in retention of exon 20. The T→C transition in the donor splice site of intron 20 in the mutant allele results in removal of introns 19 and 20 and exon 20. The base change in the donor splice site is underlined.
Figure 3
G→C transversion, which prevents phosphorylation. a, Sequence analysis of exon 19 of IKAP cDNA. cDNA from a normal individual (DY491) and from a carrier of the minor 2 haplotype (DY374; cell line generated in our laboratory) was sequenced from PCR fragments spanning exon 19. b, Immunoprecipitation of [35S]-methionine–labeled versus [32P]-orthophosphate–labeled IKAP. The DY491 and DY374 cell lines were labeled for 6 h, with either [35S]-methionine or [32P]-orthophosphate. Whole-cell extracts were prepared from these cells, and equal amounts of acid-precipitable radioactivity were immunoprecipitated with an IKAP polyclonal antibody, as described elsewhere (Rubin et al. 1988). The levels of radiolabeled IKAP protein precipitated from [35S]-methionine–labeled and [32P]-orthophosphate–labeled DY374 cells are depicted, in the bar graph, as a percentage of the radiolabeled protein precipitated from DY491 cells. The SD for four experiments is indicated. Above the bars is an autoradiograph depicting the immunoprecipitated [35S]-labeled IKAP from DY491 (lane a) and DY374 (lane b) cells and the immunoprecipitated [32P]-labeled IKAP from DY491 (lane c) and DY374 (lane d) cells. Immunoprecipitations of IKAP from three other non-FD cell lines yielded results similar to that observed with the DY491 cell line (data not shown).
Figure 4
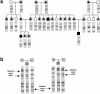
Genotype analysis, using SSCP, of FD alleles. a, PCR of the FD major allele in an extended family. Fragments spanning the intron 20 donor splice site were amplified from DNA purified from blood by use of primers 5′-GAGAACAACAAGATTCTGC-3′ and 5′-AGTCGCAAACAGTACAATGG-3′ in the presence of α[33P]-dATP. The amplified products were denatured and fractionated on a nondenaturing 5% acrylamide gel at 4°C. Circles denote females, and squares denote males. All-white symbols represent carriers of normal alleles; symbols containing black dots represent heterozygous carriers of the major FD haplotype; blackened symbols represent FD-affected homozygous individuals with two mutated alleles. b, Multiplex PCR of FD major and FD minor 2 alleles in two families. PCR was performed as in panel a, except that two additional primers—5′-GCAGTTAATGGAGAGTGGCT-3′ and 5′-ATGCTTGGTACTTGGCTG-3′—which amplify exon 19, were included in the reactions. PCR reactions were analyzed as described above. Parental carriers of the major or minor 2 allele are denoted by horizontal lines or vertical lines, respectively. FD-affected offspring with both major and minor 2 alleles are denoted by cross-hatching. Haplotype analyses were performed with the polymorphic markers 164D1, D9S1677, and 157A3, as described elsewhere (Blumenfeld et al. 1999). All blood samples were obtained with informed patient consent.
Figure 5
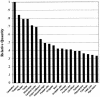
Differential expression of IKAP, by tissue type. A Multiple Tissue Expression Array (Clontech) containing RNA from 76 different human tissues and developmental stages was probed with a radiolabeled 556-bp cDNA fragment spanning exons 23–27 of IKAP and was washed according to the manufacturer’s instructions. The probed array was subjected to autoradiography and densitometric scanning, to quantitate relative levels of tissue expression. The 20 tissues that showed the highest level of expression are depicted. The highest level of expression was observed in the cerebellum, whose level was set at 1.0; the relative expression levels in the other 19 tissues are shown. The amounts of poly A+ RNA in the tissue samples on the array have been normalized on the basis of eight housekeeping genes.
Similar articles
- Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia.
Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, Liebert CB, Chadwick B, Idelson M, Reznik L, Robbins C, Makalowska I, Brownstein M, Krappmann D, Scheidereit C, Maayan C, Axelrod FB, Gusella JF. Slaugenhaupt SA, et al. Am J Hum Genet. 2001 Mar;68(3):598-605. doi: 10.1086/318810. Epub 2001 Jan 22. Am J Hum Genet. 2001. PMID: 11179008 Free PMC article. - Genomic organization and chromosomal localization of the mouse IKBKAP gene.
Coli R, Anderson SL, Volpi SA, Rubin BY. Coli R, et al. Gene. 2001 Nov 14;279(1):81-9. doi: 10.1016/s0378-1119(01)00737-5. Gene. 2001. PMID: 11722848 - Tocotrienols induce IKBKAP expression: a possible therapy for familial dysautonomia.
Anderson SL, Qiu J, Rubin BY. Anderson SL, et al. Biochem Biophys Res Commun. 2003 Jun 20;306(1):303-9. doi: 10.1016/s0006-291x(03)00971-9. Biochem Biophys Res Commun. 2003. PMID: 12788105 - Familial dysautonomia.
Slaugenhaupt SA, Gusella JF. Slaugenhaupt SA, et al. Curr Opin Genet Dev. 2002 Jun;12(3):307-11. doi: 10.1016/s0959-437x(02)00303-9. Curr Opin Genet Dev. 2002. PMID: 12076674 Review. - The molecular basis of familial dysautonomia: overview, new discoveries and implications for directed therapies.
Rubin BY, Anderson SL. Rubin BY, et al. Neuromolecular Med. 2008;10(3):148-56. doi: 10.1007/s12017-007-8019-5. Epub 2007 Nov 6. Neuromolecular Med. 2008. PMID: 17985250 Review.
Cited by
- The role of structure in regulatory RNA elements.
Tants JN, Schlundt A. Tants JN, et al. Biosci Rep. 2024 Oct 30;44(10):BSR20240139. doi: 10.1042/BSR20240139. Biosci Rep. 2024. PMID: 39364891 Free PMC article. Review. - ELP1, the Gene Mutated in Familial Dysautonomia, Is Required for Normal Enteric Nervous System Development and Maintenance and for Gut Epithelium Homeostasis.
Chaverra M, Cheney AM, Scheel A, Miller A, George L, Schultz A, Henningsen K, Kominsky D, Walk H, Kennedy WR, Kaufmann H, Walk S, Copié V, Lefcort F. Chaverra M, et al. J Neurosci. 2024 Sep 11;44(37):e2253232024. doi: 10.1523/JNEUROSCI.2253-23.2024. J Neurosci. 2024. PMID: 39138000 - A Closer Look at Familial Dysautonomia from a Social Communication Perspective: A Case Report and Review of Literature.
Turan B, Yitik Tonkaz G, Selçuk Esin İ, Burak Dursun O. Turan B, et al. Psychiatry Clin Psychopharmacol. 2022 Jun 1;32(2):178-180. doi: 10.5152/pcp.2022.21210. eCollection 2022 Jun. Psychiatry Clin Psychopharmacol. 2022. PMID: 38764866 Free PMC article. - The Relationship Between Scoliosis, Spinal Bone Density, and Truncal Muscle Strength in Familial Dysautonomia Patients.
Yovchev I, Maayan C, Simanovsky N, Foldes AJ, Brooks R, Kaplan L, Meiner Z, Cheishvili D. Yovchev I, et al. Calcif Tissue Int. 2024 Mar;114(3):222-227. doi: 10.1007/s00223-023-01164-2. Epub 2023 Nov 29. Calcif Tissue Int. 2024. PMID: 38030713 - Reduction of retinal ganglion cell death in mouse models of familial dysautonomia using AAV-mediated gene therapy and splicing modulators.
Schultz A, Cheng SY, Kirchner E, Costello S, Miettinen H, Chaverra M, King C, George L, Zhao X, Narasimhan J, Weetall M, Slaugenhaupt S, Morini E, Punzo C, Lefcort F. Schultz A, et al. Sci Rep. 2023 Oct 30;13(1):18600. doi: 10.1038/s41598-023-45376-w. Sci Rep. 2023. PMID: 37903840 Free PMC article.
References
Electronic-Database Information
- Genbank, http://www.ncbi.nlm.nih.gov/Genbank/ (for IKAP cDNA [accession number NM_003640])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for hereditary sensory neuropathy type III [MIM 223900])
References
- Axelrod FB (1996) Familial dysautonomia. In: Robertson D, Low PA, Polinsky RJ (eds) Primer on the autonomic nervous system. Academic Press, San Diego, pp 242–249
- Axelrod FB, Nachtigal R, Dancis J (1974) Familial dysautonomia: diagnosis, pathogenesis and management. Adv Pediatr 21:75–96 - PubMed
- Blumenfeld A, Slaugenhaupt SA, Liebert CB, Temper V, Maayan C, Gill S, Lucente DE, Idelson M, MacCormack K, Monahan MA, Mull J, Leyne M, Mendillo M, Schiripo T, Mishori E, Breakefield X, Axelrod FB, Gusella JF (1999) Precise genetic mapping and haplotype analysis of the familial dysautonomia gene on human chromosome 9q31. Am J Hum Genet 64:1110–1118 - PMC - PubMed
- Breakefield XO, Ozelius L, Bothwell MA, Chao MV, Axelrod F, Kramer PL, Kidd KK, et al (1986) DNA polymorphisms for the nerve growth factor receptor gene exclude its role in familial dysautonomia. Mol Biol Med 3:483–94 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials