The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts - PubMed (original) (raw)
The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts
W Wang et al. EMBO J. 2001.
Abstract
The yeast UPF1, UPF2 and UPF3 genes encode trans-acting factors of the nonsense-mediated mRNA decay pathway. In addition, the upf1Delta strain demonstrates a nonsense suppression phenotype and Upf1p has been shown to interact with the release factors eRF1 and eRF3. In this report, we show that both upf2Delta and upf3Delta strains demonstrate a nonsense suppression phenotype independent of their effect on mRNA turnover. We also demonstrate that Upf2p and Upf3p interact with eRF3, and that their ability to bind eRF3 correlates with their ability to complement the nonsense suppression phenotype. In vitro experiments demonstrate that Upf2p, Upf3p and eRF1 compete with each other for interacting with eRF3. Con versely, Upf1p binds to a different region of eRF3 and can form a complex with these factors. These results suggest a sequential surveillance complex assembly pathway, which occurs during the premature translation termination process. We propose that the observed nonsense suppression phenotype in the upfDelta strains can be attributed to a defect in the surveillance complex assembly.
Figures
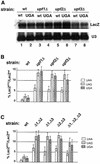
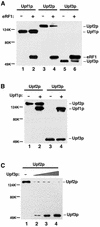
Fig. 1. The upf2Δ and upf3Δ strains demonstrate a nonsense suppression phenotype. The UPF1, UPF2 and UPF3 genes in a wild-type yeast strain (KC2) were disrupted individually. Ten-fold serial dilutions of mid-log phase cells were plated on synthetic complete (SC) (right panel) and –tyr (left panel) plates, and their growth at 30°C was monitored.
Fig. 2. (A) The abundance of wild-type and UGA-containing LacZ transcripts in the wild-type and upfΔ strains. Total RNAs isolated from the specified strains were separated on a 1% agarose gel and probed with 32P-labeled LacZ and U3 probes. (B) The nonsense suppression activity in wild-type and upfΔ strains was assessed quantitatively using a β-gal reporter system. The indicated yeast strains were transformed with either a wild-type LacZ gene or a LacZ gene containing the specified nonsense codon. The assays were performed as described in Materials and methods. (C) Nonsense suppression activity in wild type and strains harboring multiple UPF deletions.
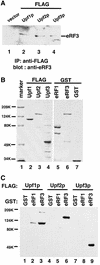
Fig. 3. Upf2p and Upf3p interact with the release factor eRF3. (A) Co-immunoprecipitation. Cells were transformed with either vector alone (lane 1) or vector expressing FLAG-tagged Upf1p, Upf2p or Upf3p (lanes 2–4). Cytoplasmic extracts were prepared and immuno precipitated with an anti-FLAG antibody. The immunoprecipitates were resolved by SDS–PAGE and subjected to immunoblotting with an anti-eRF3 polyclonal antibody. (B) Coomassie Blue staining of the purified fusion proteins. The protein molecular weight markers are as indicated. (C) GST pull-down experiment. Purified GST, GST–eRF1 or GST–eRF3 (1.0 µg each) was combined with glutathione–Sepharose beads and FLAG-Upf1p (1.0 µg), Upf2p (1.0 µg) or Upf3p (0.5 µg). Following incubation and extensive washing, the proteins remaining associated with the beads were analyzed by SDS–PAGE and immunoblotting with an anti-FLAG antibody.
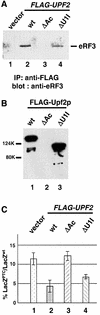
Fig. 4. The ability of Upf2p to complement the nonsense suppression phenotype in a upf2Δ strain correlates with its ability to interact with eRF3. (A) Co-immunoprecipitation. Cells were transformed with either vector alone or vector expressing the specified FLAG-Upf2p. Cytoplasmic extracts were prepared and immunoprecipitated with an anti-FLAG antibody. The immunoprecipitates were separated by SDS–PAGE and immunoblotted using the anti-eRF3 antibody. (B) GST pull-down experiment. Purified wild-type and mutant FLAG-Upf2 proteins (1.0 µg each) were combined with purified GST–eRF3 (1.0 µg) and glutathione–Sepharose beads. Following incubation and extensive washing, the proteins remaining associated with the beads were separated by SDS–PAGE and detected by the anti-FLAG antibody. (C) The ability of upf2 mutants to complement the nonsense suppression phenotype in a upf2Δ strain. Cells harboring either wild type or a UGA-containing LacZ gene were transformed with either vector alone or the vector expressing the specified upf2 gene. The assays were performed as described in Materials and methods.
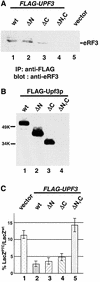
Fig. 5. The ability of Upf3p to complement the nonsense suppression phenotype in a upf3Δ strain correlates with its ability to interact with eRF3. (A) Co-immunoprecipitation. Cells were transformed with either vector alone (lane 5) or a vector expressing the specified FLAG-Upf3p. Cytoplasmic extracts were prepared and immunoprecipitated with an anti-FLAG antibody, and the immunoprecipitates were subjected to SDS–PAGE and immunoblotted by the anti-eRF3 antibody. (B) GST pull-down experiment. Purified wild-type and mutant FLAG-Upf3 proteins (0.5 µg each) were combined with purified GST–eRF3 (1.0 µg) and glutathione–Sepharose beads. Following incubation and extensive washing, the proteins remaining associated with the beads were separated by SDS–PAGE and detected by the anti-FLAG antibody. (C) The ability of upf3 mutants to complement the nonsense suppression phenotype in a upf3Δ strain. Cells harboring either wild-type or a UGA-containing LacZ gene were transformed with either the vector expressing the specified upf3 gene (lanes 1–4) or vector alone (lane 5). The assays were performed as described in Materials and methods.
Fig. 6. The Upf proteins and eRF1 interact with the essential GTPase domain of eRF3. (A) Schematic diagram of the domain structure of the yeast release factor eRF3 (Sup35p). (B) GST pull-down experiment. Purified GST–eRF3 (1.0 µg, lanes 1–4), eRF3-N254Δ (1.0 µg, lanes 5–8) or eRF3-N465 (1.0 µg, lanes 9–12) was combined with FLAG-Upf1p (1.0 µg, lanes 1, 5 and 9), -Upf2p (1.0 µg, lanes 2, 6 and 10), -Upf3p (0.5 µg, lanes 3, 7 and 11) or -eRF1 (0.5 µg, lanes 4, 8 and 12). Following incubation and extensive washing, the proteins remaining associated with the beads were resolved on 12% SDS–PAGE and detected by the anti-FLAG antibody.
Fig. 7. Upf2p, Upf3p and eRF1 competed with each other, but not with Upf1p, for binding to eRF3. (A) Analysis of the interactions between FLAG-Upf1p (1.0 µg), -Upf2p (1.0 µg), -Upf3p (0.5 µg) and GST–eRF3 (0.5 µg) in the absence or presence of FLAG-eRF1 (0.5 µg) by GST pull-down experiments. (B) Analysis of the interactions between FLAG-Upf2p (1.0 µg), -Upf3p (0.5 µg) and GST–eRF3 (0.5 µg) in the absence or presence of FLAG-Upf1p (1.0 µg) by GST pull-down experiments. (C) Analysis of the interactions between FLAG-Upf2p (1.0 µg) and GST–eRF3 (0.5 µg) in the absence (lane 1) or presence of increasing amounts of FLAG-Upf3p (lanes 2–4, 0.5, 1.0 and 2.0 µg) by GST pull-down experiments.
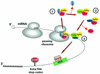
Fig. 8. Model for the sequential surveillance complex assembly pathway. (1) The translating ribosome pauses at a premature termination codon and signals the eRF1–eRF3 complex to bind to its A site. The Upf1p becomes associated with the eRF1–eRF3 complex during the termination process. (2) After hydrolysis of the peptidyl-tRNA bond, eRF1 dissociates from the ribosome. Dissociation of eRF1 allows either Upf2p or Upf3p to bind the eRF3–Upf1p complex. (3) Rearrangement of the complex: Upf3p (or Upf2p) joins the complex and displaces eRF3 to form the mature surveillance complex.
Similar articles
- Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover.
Weng Y, Czaplinski K, Peltz SW. Weng Y, et al. Mol Cell Biol. 1996 Oct;16(10):5491-506. doi: 10.1128/MCB.16.10.5491. Mol Cell Biol. 1996. PMID: 8816462 Free PMC article. - Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p.
Maderazo AB, He F, Mangus DA, Jacobson A. Maderazo AB, et al. Mol Cell Biol. 2000 Jul;20(13):4591-603. doi: 10.1128/MCB.20.13.4591-4603.2000. Mol Cell Biol. 2000. PMID: 10848586 Free PMC article. - Overexpression of Upf1p compensates for mitochondrial splicing deficiency independently of its role in mRNA surveillance.
de Pinto B, Lippolis R, Castaldo R, Altamura N. de Pinto B, et al. Mol Microbiol. 2004 Feb;51(4):1129-42. doi: 10.1046/j.1365-2958.2003.03889.x. Mol Microbiol. 2004. PMID: 14763985 - NMD: At the crossroads between translation termination and ribosome recycling.
Celik A, Kervestin S, Jacobson A. Celik A, et al. Biochimie. 2015 Jul;114:2-9. doi: 10.1016/j.biochi.2014.10.027. Epub 2014 Nov 13. Biochimie. 2015. PMID: 25446649 Free PMC article. Review. - Transcript selection and the recruitment of mRNA decay factors for NMD in Saccharomyces cerevisiae.
Culbertson MR, Neeno-Eckwall E. Culbertson MR, et al. RNA. 2005 Sep;11(9):1333-9. doi: 10.1261/rna.2113605. Epub 2005 Jul 25. RNA. 2005. PMID: 16043493 Free PMC article. Review.
Cited by
- Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes.
Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Fleischer TC, et al. Genes Dev. 2006 May 15;20(10):1294-307. doi: 10.1101/gad.1422006. Genes Dev. 2006. PMID: 16702403 Free PMC article. - Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay.
Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Kashima I, et al. Genes Dev. 2006 Feb 1;20(3):355-67. doi: 10.1101/gad.1389006. Genes Dev. 2006. PMID: 16452507 Free PMC article. - Gene overexpression as a tool for identifying new trans-acting factors involved in translation termination in Saccharomyces cerevisiae.
Namy O, Hatin I, Stahl G, Liu H, Barnay S, Bidou L, Rousset JP. Namy O, et al. Genetics. 2002 Jun;161(2):585-94. doi: 10.1093/genetics/161.2.585. Genetics. 2002. PMID: 12072456 Free PMC article. - Multiple transcripts from a 3'-UTR reporter vary in sensitivity to nonsense-mediated mRNA decay in Saccharomyces cerevisiae.
Zaborske JM, Zeitler B, Culbertson MR. Zaborske JM, et al. PLoS One. 2013 Nov 18;8(11):e80981. doi: 10.1371/journal.pone.0080981. eCollection 2013. PLoS One. 2013. PMID: 24260526 Free PMC article. - An RNA decay factor wears a new coat: UPF3B modulates translation termination.
Gao Z, Wilkinson M. Gao Z, et al. F1000Res. 2017 Dec 20;6:2159. doi: 10.12688/f1000research.12704.1. eCollection 2017. F1000Res. 2017. PMID: 29333258 Free PMC article. Review.
References
- Atkin A.L., Schenkman,L.R., Eastham,M., Dahlseid,J.N., Lelivelt,M.J. and Culbertson,M.R. (1997) Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem., 272, 22163–22172. - PubMed
- Cui Y., Hagan,K.W., Zhang,S. and Peltz,S.W. (1995) Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev., 9, 423–436. - PubMed
- Czaplinski K., Ruiz-Echevarria,M.J., Paushkin,S.V., Han,X., Weng,Y., Perlick,H.A., Dietz,H.C., Ter-Avanesyan,M.D. and Peltz,S.W. (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev., 12, 1665–1677. - PMC - PubMed
- Czaplinski K., Ruiz-Echevarria,M.J., Gonzales,C.I. and Peltz,S.W. (1999) Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. BioEssays, 21, 685–696. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases