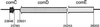Natural genetic transformation of Streptococcus mutans growing in biofilms - PubMed (original) (raw)
Natural genetic transformation of Streptococcus mutans growing in biofilms
Y H Li et al. J Bacteriol. 2001 Feb.
Abstract
Streptococcus mutans is a bacterium that has evolved to be dependent upon a biofilm "lifestyle" for survival and persistence in its natural ecosystem, dental plaque. We initiated this study to identify the genes involved in the development of genetic competence in S. mutans and to assay the natural genetic transformability of biofilm-grown cells. Using genomic analyses, we identified a quorum-sensing peptide pheromone signaling system similar to those previously found in other streptococci. The genetic locus of this system comprises three genes, comC, comD, and comE, that encode a precursor to the peptide competence factor, a histidine kinase, and a response regulator, respectively. We deduced the sequence of comC and its active pheromone product and chemically synthesized the corresponding 21-amino-acid competence-stimulating peptide (CSP). Addition of CSP to noncompetent cells facilitated increased transformation frequencies, with typically 1% of the total cell population transformed. To further confirm the roles of these genes in genetic competence, we inactivated them by insertion-duplication mutagenesis or allelic replacement followed by assays of transformation efficiency. We also demonstrated that biofilm-grown S. mutans cells were transformed at a rate 10- to 600-fold higher than planktonic S. mutans cells. Donor DNA included a suicide plasmid, S. mutans chromosomal DNA harboring a heterologous erythromycin resistance gene, and a replicative plasmid. The cells were optimally transformed during the formation of 8- to 16-h-old biofilms primarily consisting of microcolonies on solid surfaces. We also found that dead cells in the biofilms could act as donors of a chromosomally encoded antibiotic resistance determinant. This work demonstrated that a peptide pheromone system controls genetic competence in S. mutans and that the system functions optimally when the cells are living in actively growing biofilms.
Figures
FIG. 1
Deduced amino acid sequences of CSPs from various S. mutans strains. Boldface type indicates variant amino acids. ^, predicted cleavage site.
FIG. 2
Orientations and locations of the comC, comD, and comE genes in the partially completed S. mutans genome database, available from the OU-ACGT website. The numbers correspond to the base pair designations as they appear in contig 459, file date 12 November 1999.
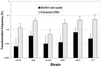
FIG. 3
Natural genetic transformation of six S. mutans strains with saturating concentrations of integration plasmid pVA-GTFA as donor DNA (1 μg/ml). Biofilms accumulated on the surfaces of polystyrene microtiter plates for 20 h. Biofilm-grown cells of all strains were able to incorporate foreign DNA more efficiently (10- to 600-fold) than their planktonic (Plank.) counterparts. The transformation frequency is expressed as the percentage of viable cells transformed to erythromycin resistance. The results are expressed as the mean + standard deviation of four independent cultures.
FIG. 4
Kinetics of biofilm formation of S. mutans strain UA159 on polystyrene microtiter plates (A) and on glass rods suspended in the chemostat (B). The inset in panel A shows the mean viable biofilm cell counts of S. mutans strains during 20 h of accumulation. Biofilm formation by S. mutans grown in BM usually showed three accumulation phases: (i) the adherence phase (0 to 4 h), (ii) the active accumulation phase (4 to 20 h), and (iii) the slow or plateau accumulation phase (after 20 h). Results are expressed as mean CFU ± standard deviation (SD) of four independent cultures for polystyrene-grown biofilms and of three rods from each of two independent cultures for fermentor-grown biofilms.
FIG. 5
Scanning electron micrographs of biofilms accumulated on polystyrene microtiter wells at various times.
FIG. 6
Natural transformation with heat-killed biofilms of strain YD025 (strain UA159 harboring chromosomally integrated pVA-GTFA; Emr) as a source of donor DNA (lysate). Extracted chromosomal DNA (10 μg/ml) from the same strain was used as a control. Results are expressed as the mean + standard deviation of three independent experiments.
FIG. 7
Effect of dilution rate on competence development of S. mutans UA159 grown in continuous cultures. Twenty-hour biofilms were assayed for transformation with pVA-GTFA as the donor DNA (1 μg/ml). Plank., planktonic.
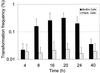
FIG. 8
Time course experiment of natural transformation of S. mutans UA159 grown at a D of 0.5 h−1, pVA-GTFA was used as the donor DNA (1 μg/ml). Plank., planktonic.
Similar articles
- ComX activity of Streptococcus mutans growing in biofilms.
Aspiras MB, Ellen RP, Cvitkovitch DG. Aspiras MB, et al. FEMS Microbiol Lett. 2004 Sep 1;238(1):167-74. doi: 10.1016/j.femsle.2004.07.032. FEMS Microbiol Lett. 2004. PMID: 15336418 - Inhibiting effects of Streptococcus salivarius on competence-stimulating peptide-dependent biofilm formation by Streptococcus mutans.
Tamura S, Yonezawa H, Motegi M, Nakao R, Yoneda S, Watanabe H, Yamazaki T, Senpuku H. Tamura S, et al. Oral Microbiol Immunol. 2009 Apr;24(2):152-61. doi: 10.1111/j.1399-302X.2008.00489.x. Oral Microbiol Immunol. 2009. PMID: 19239643 - Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms.
Li YH, Hanna MN, Svensäter G, Ellen RP, Cvitkovitch DG. Li YH, et al. J Bacteriol. 2001 Dec;183(23):6875-84. doi: 10.1128/JB.183.23.6875-6884.2001. J Bacteriol. 2001. PMID: 11698377 Free PMC article. - Quorum sensing and biofilm formation by Streptococcus mutans.
Senadheera D, Cvitkovitch DG. Senadheera D, et al. Adv Exp Med Biol. 2008;631:178-88. doi: 10.1007/978-0-387-78885-2_12. Adv Exp Med Biol. 2008. PMID: 18792689 Review. - Genetic competence and transformation in oral streptococci.
Cvitkovitch DG. Cvitkovitch DG. Crit Rev Oral Biol Med. 2001;12(3):217-43. doi: 10.1177/10454411010120030201. Crit Rev Oral Biol Med. 2001. PMID: 11497374 Review.
Cited by
- Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX.
Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ. Son M, et al. Mol Microbiol. 2012 Oct;86(2):258-72. doi: 10.1111/j.1365-2958.2012.08187.x. Epub 2012 Aug 16. Mol Microbiol. 2012. PMID: 22845615 Free PMC article. - One if by land, two if by sea: signalling to the ranks with CSP and XIP.
Federle MJ, Morrison DA. Federle MJ, et al. Mol Microbiol. 2012 Oct;86(2):241-5. doi: 10.1111/mmi.12029. Epub 2012 Sep 19. Mol Microbiol. 2012. PMID: 22958130 Free PMC article. - Identification of the Streptococcus mutans LytST two-component regulon reveals its contribution to oxidative stress tolerance.
Ahn SJ, Qu MD, Roberts E, Burne RA, Rice KC. Ahn SJ, et al. BMC Microbiol. 2012 Sep 1;12:187. doi: 10.1186/1471-2180-12-187. BMC Microbiol. 2012. PMID: 22937869 Free PMC article. - Membrane composition changes and physiological adaptation by Streptococcus mutans signal recognition particle pathway mutants.
Hasona A, Zuobi-Hasona K, Crowley PJ, Abranches J, Ruelf MA, Bleiweis AS, Brady LJ. Hasona A, et al. J Bacteriol. 2007 Feb;189(4):1219-30. doi: 10.1128/JB.01146-06. Epub 2006 Nov 3. J Bacteriol. 2007. PMID: 17085548 Free PMC article. - Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms.
Michaelis C, Grohmann E. Michaelis C, et al. Antibiotics (Basel). 2023 Feb 4;12(2):328. doi: 10.3390/antibiotics12020328. Antibiotics (Basel). 2023. PMID: 36830238 Free PMC article. Review.
References
- Bowden G H, Hamilton I R. Survival of oral bacteria. Crit Rev Oral Biol Med. 1998;9:54–85. - PubMed
- Bradshaw D J, Homer K A, Marsh P D, Beighton D. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology. 1994;140:3407–3412. - PubMed
- Burne R A, Rubinfeld B, Bowen W H, Yasbin R E. Tight genetic linkage of a glucosyltransferase and dextranase of Streptococcus mutans GS-5. J Dent Res. 1986;65:1392–1401. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources