Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli - PubMed (original) (raw)
Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli
D Scholl et al. J Virol. 2001 Mar.
Abstract
A virulent double-stranded DNA bacteriophage, Phi K1-5, has been isolated and found to be capable of infecting Escherichia coli strains that possess either the K1 or the K5 polysaccharide capsule. Electron micrographs show that the virion consists of a small icosohedral head with short tail spikes, similar to members of the Podoviridae family. DNA sequence analysis of the region encoding the tail fiber protein showed two open reading frames encoding previously characterized hydrolytic phage tail fiber proteins. The first is the K5 lyase protein gene of Phi K5, which allows this phage to specifically infect K5 E. coli strains. A second open reading frame encodes a protein almost identical in amino acid sequence to the N-acetylneuraminidase (endosialidase) protein of Phi K1E, which allows this phage to specifically infect K1 strains of E. coli. We provide experimental evidence that mature phage particles contain both tail fiber proteins, and mutational analysis indicates that each protein can be independently inactivated. A comparison of the tail gene regions of Phi K5, Phi K1E, and Phi K1-5 shows that the genes are arranged in a modular or cassette configuration and suggests that this family of phages can broaden host range by horizontal gene transfer.
Figures
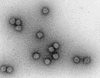
FIG. 1
Electron micrograph of ΦK1-5 negatively stained with phosphotungstic acid at a magnification of ×115,500. Morphologically this phage can be classified in the Podoviridae family which includes T7 and SP6.
FIG. 2
Comparison of the coding regions of the tail proteins of ΦK1-5, ΦK5, and ΦK1E. All three phages share sequence similarity in the upstream region (which contains an SP6 promoter) as well as an 85-base intergenic region. Just downstream of the promoter, ΦK1-5 and ΦK5 encode a lyase protein and ΦK1E encodes ORFL. Immediately following the termination codons of the lyases or ORFL is the intergenic region that contains a potential hairpin structure, the first of which could be a Rho-independent transcription terminator. Immediately following this, ΦK1-5 and ΦK1E encode an endosialidase where ΦK5 encodes ORFP. None of the three phages have any coding regions downstream, and the DNA molecule ends in all three cases. No homology exists in this terminal region.
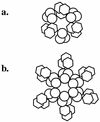
FIG. 3
Two possible models for the arrangement of the tails proteins on the phage capsid. (a) There are three copies of each tail forming a hexamer. (b) There are six copies of each tail. One is attached to the head and is part of the core of the tail; the other is then attached to the first tail protein, in effect making a longer tail fiber with two different enzymatic activities.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Far Posterior Approach for Rib Fracture Fixation: Surgical Technique and Tips.
Manes TJ, DeGenova DT, Taylor BC, Patel JN. Manes TJ, et al. JBJS Essent Surg Tech. 2024 Dec 6;14(4):e23.00094. doi: 10.2106/JBJS.ST.23.00094. eCollection 2024 Oct-Dec. JBJS Essent Surg Tech. 2024. PMID: 39650795 Free PMC article. - Antiviral function and viral antagonism of the rapidly evolving dynein activating adaptor NINL.
Stevens DA, Beierschmitt C, Mahesula S, Corley MR, Salogiannis J, Tsu BV, Cao B, Ryan AP, Hakozawki H, Reck-Peterson SL, Daugherty MD. Stevens DA, et al. Elife. 2022 Oct 12;11:e81606. doi: 10.7554/eLife.81606. Elife. 2022. PMID: 36222652 Free PMC article. - Unlocking data: Decision-maker perspectives on cross-sectoral data sharing and linkage as part of a whole-systems approach to public health policy and practice.
Tweed E, Cimova K, Craig P, Allik M, Brown D, Campbell M, Henderson D, Mayor C, Meier P, Watson N. Tweed E, et al. Public Health Res (Southampt). 2024 Nov 20:1-30. doi: 10.3310/KYTW2173. Online ahead of print. Public Health Res (Southampt). 2024. PMID: 39582242 - Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.
Triana L, Palacios Huatuco RM, Campilgio G, Liscano E. Triana L, et al. Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
- Modular prophage interactions driven by capsule serotype select for capsule loss under phage predation.
de Sousa JAM, Buffet A, Haudiquet M, Rocha EPC, Rendueles O. de Sousa JAM, et al. ISME J. 2020 Dec;14(12):2980-2996. doi: 10.1038/s41396-020-0726-z. Epub 2020 Jul 30. ISME J. 2020. PMID: 32732904 Free PMC article. - Characterization of extended-host-range pseudo-T-even bacteriophage Kpp95 isolated on Klebsiella pneumoniae.
Wu LT, Chang SY, Yen MR, Yang TC, Tseng YH. Wu LT, et al. Appl Environ Microbiol. 2007 Apr;73(8):2532-40. doi: 10.1128/AEM.02113-06. Epub 2007 Mar 2. Appl Environ Microbiol. 2007. PMID: 17337566 Free PMC article. - Why bacteriophage encode exotoxins and other virulence factors.
Abedon ST, Lejeune JT. Abedon ST, et al. Evol Bioinform Online. 2007 Feb 28;1:97-110. Evol Bioinform Online. 2007. PMID: 19325857 Free PMC article. - Viral ecology of organic and inorganic particles in aquatic systems: avenues for further research.
Weinbauer MG, Bettarel Y, Cattaneo R, Luef B, Maier C, Motegi C, Peduzzi P, Mari X. Weinbauer MG, et al. Aquat Microb Ecol. 2009 Dec;57(3):321-341. doi: 10.3354/ame01363. Epub 2009 Nov 24. Aquat Microb Ecol. 2009. PMID: 27478304 Free PMC article. - Complete Genome Sequence of Enterotoxigenic Escherichia coli Podophage LL11.
Theodore M, Lessor L, O'Leary C, Kongari R, Gill J, Liu M. Theodore M, et al. Microbiol Resour Announc. 2019 Aug 8;8(32):e00693-19. doi: 10.1128/MRA.00693-19. Microbiol Resour Announc. 2019. PMID: 31395642 Free PMC article.
References
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Botstein D. A theory of modular evolution for bacteriophages. Ann N Y Acad Sci. 1980;354:484–490. - PubMed
- Campbell A, Botstein D. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 365–380.
- Chandry P S, Moore S C, Boyce J D, Davidson B E, Hillier A J. Analysis of the DNA sequence, gene expression, origin of replication, and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol Microbiol. 1997;26:49–64. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials