Evolutionary relationships among parvoviruses: virus-host coevolution among autonomous primate parvoviruses and links between adeno-associated and avian parvoviruses - PubMed (original) (raw)
Evolutionary relationships among parvoviruses: virus-host coevolution among autonomous primate parvoviruses and links between adeno-associated and avian parvoviruses
V V Lukashov et al. J Virol. 2001 Mar.
Abstract
The current classification of parvoviruses is based on virus host range and helper virus dependence, while little data on evolutionary relationships among viruses are available. We identified and analyzed 472 sequences of parvoviruses, among which there were (virtually) full-length genomes of all 41 viruses currently recognized as individual species within the family Parvoviridae. Our phylogenetic analysis of full-length genomes as well as open reading frames distinguished three evolutionary groups of parvoviruses from vertebrates: (i) the human helper-dependent adeno-associated virus (AAV) serotypes 1 to 6 and the autonomous avian parvoviruses; (ii) the bovine, chipmunk, and autonomous primate parvoviruses, including human viruses B19 and V9; and (iii) the parvoviruses from rodents (except for chipmunks), carnivores, and pigs. Each of these three evolutionary groups could be further subdivided, reflecting both virus-host coevolution and multiple cross-species transmissions in the evolutionary history of parvoviruses. No parvoviruses from invertebrates clustered with vertebrate parvoviruses. Our analysis provided evidence for negative selection among parvoviruses, the independent evolution of their genes, and recombination among parvoviruses from rodents. The topology of the phylogenetic tree of autonomous human and simian parvoviruses matched exactly the topology of the primate family tree, as based on the analysis of primate mitochondrial DNA. Viruses belonging to the AAV group were not evolutionarily linked to other primate parvoviruses but were linked to the parvoviruses of birds. The two lineages of human parvoviruses may have resulted from independent ancient zoonotic infections. Our results provide an argument for reclassification of Parvovirinae based on evolutionary relationships among viruses.
Figures
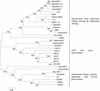
FIG. 1
The three evolutionary groups of Parvovirinae. The neighbor-joining phylogenetic tree is based on the analysis of (virtually) full-length genomes of all members of the Parvovirinae subfamily that are recognized as individual virus species, one sequence per species (except for the B19 virus, for which a consensus, ConsB19, of 215 available sequences is also included). For RaccoonPV, only a shorter sequence is available. Bootstrap values are shown (100 replications). Sequences used in this analysis are in boldface in Table 1. For virus abbreviations, see Table 1.
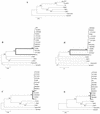
FIG. 2
Phylogenetic relationships among the AAV serotypes 1 to 6 and parvoviruses from GPV, Barbarie duck parvovirus (BarbduckPV), and MDPV (MuscduckPV). Bootstrap values are shown (100 replications). (A) Relationships based on nucleotide p distances among full-length genome sequences; (B to D) relationships based on nucleotide Kimura two-parameter distances, Ds, and Da, respectively, for orf1; (E to G) nucleotide distances, Ds, and Da, respectively, for orf2. For panels B and E, positions of AAV-2 and AAV-4 are marked. Virus abbreviations are listed in Table 1.
FIG. 3
Phylogenetic relationships among the autonomous primate, chipmunk, and bovine parvoviruses. In addition to sequences used in Fig. 1, 11 more sequences of B19 are included (labeled by their GenBank accession numbers). Bootstrap values above 70 are shown (100 replications). (A) Relationships based on nucleotide p distances among full-length genome sequences; (B and C) relationships based on Ds and Da, respectively, for orf1; (D and E) relationships based on Ds and Da, respectively, for orf2. Branches between the B19 cluster and V9 are in boldface (B to E). Virus abbreviations are in Table 1.
FIG. 4
Phylogenetic relationships among parvoviruses from rodents, carnivores, and pigs. The four phylogenetic subgroups are shown. (A) All full-length sequences available for each virus species are included (labeled by their virus names and the GenBank accession numbers). Bootstrap values above 70 are shown (100 replications). (A) relationships based on nucleotide p distances among full-length genome sequences; (B to D) relationships based on the nucleotide Kimura two-parameter distances, Ds, and Da, respectively, for orf1; (E to G) relationships based on nucleotide Kimura two-parameter distances, Ds, and Da, respectively, for orf2. In orf1 (B to D), MousePV, HamsterPV, and MVM form a homogeneous cluster (B, grey box), to which LuIII (arrow) is an outlier. In contrast, in orf2 (E to G), MousePV and HamsterPV (E, open box) cluster with LuIII (arrow) and not with MVM (grey box). Virus abbreviations are in Table 1.
FIG. 4
Phylogenetic relationships among parvoviruses from rodents, carnivores, and pigs. The four phylogenetic subgroups are shown. (A) All full-length sequences available for each virus species are included (labeled by their virus names and the GenBank accession numbers). Bootstrap values above 70 are shown (100 replications). (A) relationships based on nucleotide p distances among full-length genome sequences; (B to D) relationships based on the nucleotide Kimura two-parameter distances, Ds, and Da, respectively, for orf1; (E to G) relationships based on nucleotide Kimura two-parameter distances, Ds, and Da, respectively, for orf2. In orf1 (B to D), MousePV, HamsterPV, and MVM form a homogeneous cluster (B, grey box), to which LuIII (arrow) is an outlier. In contrast, in orf2 (E to G), MousePV and HamsterPV (E, open box) cluster with LuIII (arrow) and not with MVM (grey box). Virus abbreviations are in Table 1.
FIG. 4
Phylogenetic relationships among parvoviruses from rodents, carnivores, and pigs. The four phylogenetic subgroups are shown. (A) All full-length sequences available for each virus species are included (labeled by their virus names and the GenBank accession numbers). Bootstrap values above 70 are shown (100 replications). (A) relationships based on nucleotide p distances among full-length genome sequences; (B to D) relationships based on the nucleotide Kimura two-parameter distances, Ds, and Da, respectively, for orf1; (E to G) relationships based on nucleotide Kimura two-parameter distances, Ds, and Da, respectively, for orf2. In orf1 (B to D), MousePV, HamsterPV, and MVM form a homogeneous cluster (B, grey box), to which LuIII (arrow) is an outlier. In contrast, in orf2 (E to G), MousePV and HamsterPV (E, open box) cluster with LuIII (arrow) and not with MVM (grey box). Virus abbreviations are in Table 1.
FIG. 5
Bootscan analysis of the phylogenetic relationships among LuIII and parvoviruses from mice and hamsters. The three MousePV and HamsterPV were used as a query sequence group in comparison to the five MVM (comparison group 1), LuIII (comparison group 2), and H1 (outgroup). Analysis settings were as follows: window size, 400 nucleotides; step 100 nucleotides; bootstrap resampling, 100; distance, Kimura two-parameter distance; transitions/transversions ratio, 2. Arrow, recombination site.
Similar articles
- Molecular evolutionary genetic analysis of emerging parvoviruses identified in pigs.
Xiao CT, Halbur PG, Opriessnig T. Xiao CT, et al. Infect Genet Evol. 2013 Jun;16:369-76. doi: 10.1016/j.meegid.2013.03.017. Epub 2013 Mar 22. Infect Genet Evol. 2013. PMID: 23523595 - CpG dinucleotide frequencies reveal the role of host methylation capabilities in parvovirus evolution.
Upadhyay M, Samal J, Kandpal M, Vasaikar S, Biswas B, Gomes J, Vivekanandan P. Upadhyay M, et al. J Virol. 2013 Dec;87(24):13816-24. doi: 10.1128/JVI.02515-13. Epub 2013 Oct 9. J Virol. 2013. PMID: 24109231 Free PMC article. - Novel parvoviruses in reptiles and genome sequence of a lizard parvovirus shed light on Dependoparvovirus genus evolution.
Pénzes JJ, Pham HT, Benkö M, Tijssen P. Pénzes JJ, et al. J Gen Virol. 2015 Sep;96(9):2769-2779. doi: 10.1099/vir.0.000215. Epub 2015 Jun 11. J Gen Virol. 2015. PMID: 26067293 - Parvovirus variation for disease: a difference with RNA viruses?
López-Bueno A, Villarreal LP, Almendral JM. López-Bueno A, et al. Curr Top Microbiol Immunol. 2006;299:349-70. doi: 10.1007/3-540-26397-7_13. Curr Top Microbiol Immunol. 2006. PMID: 16568906 Review. - Avian parvovirus: classification, phylogeny, pathogenesis and diagnosis.
Kapgate SS, Kumanan K, Vijayarani K, Barbuddhe SB. Kapgate SS, et al. Avian Pathol. 2018 Dec;47(6):536-545. doi: 10.1080/03079457.2018.1517938. Epub 2018 Sep 24. Avian Pathol. 2018. PMID: 30246559 Review.
Cited by
- Aleutian mink disease virus in furbearing mammals in Nova Scotia, Canada.
Farid AH. Farid AH. Acta Vet Scand. 2013 Feb 8;55(1):10. doi: 10.1186/1751-0147-55-10. Acta Vet Scand. 2013. PMID: 23394546 Free PMC article. - Molecular characterization of emerging chicken and turkey parvovirus variants and novel strains in Guangxi, China.
Zhang Y, Feng B, Xie Z, Zhang M, Fan Q, Deng X, Xie Z, Li M, Zeng T, Xie L, Luo S, Huang J, Wang S. Zhang Y, et al. Sci Rep. 2023 Aug 11;13(1):13083. doi: 10.1038/s41598-023-40349-5. Sci Rep. 2023. PMID: 37567941 Free PMC article. - Recombination in eukaryotic single stranded DNA viruses.
Martin DP, Biagini P, Lefeuvre P, Golden M, Roumagnac P, Varsani A. Martin DP, et al. Viruses. 2011 Sep;3(9):1699-738. doi: 10.3390/v3091699. Epub 2011 Sep 13. Viruses. 2011. PMID: 21994803 Free PMC article. Review. - Detection of parvovirus B19 in selected high-risk patient groups & their phylogenetic & selection analysis.
Vadivel K, Mageshbabu R, Sankar S, Jain A, Perumal V, Srikanth P, Ranjan GA, Nair A, Simoes EAF, Nandagopal B, Sridharan G. Vadivel K, et al. Indian J Med Res. 2018 Apr;147(4):391-399. doi: 10.4103/ijmr.IJMR_241_16. Indian J Med Res. 2018. PMID: 29998875 Free PMC article. - Cloning of an avian adeno-associated virus (AAAV) and generation of recombinant AAAV particles.
Bossis I, Chiorini JA. Bossis I, et al. J Virol. 2003 Jun;77(12):6799-810. doi: 10.1128/jvi.77.12.6799-6810.2003. J Virol. 2003. PMID: 12768000 Free PMC article.
References
- Afanasiev B N, Galyov E E, Buchatsky L P, Kozlov Y V. Nucleotide sequence and genomic organization of Aedes densonucleosis virus. Virology. 1991;185:323–336. - PubMed
- Auguste, V., A., Garbarg-Chenon, and Q. T. Nguyen. 1999. Erythrovirus and its applications. French patent WO9928439.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources