RegA proteins from phage T4 and RB69 have conserved helix-loop groove RNA binding motifs but different RNA binding specificities - PubMed (original) (raw)
RegA proteins from phage T4 and RB69 have conserved helix-loop groove RNA binding motifs but different RNA binding specificities
T K Sengupta et al. Nucleic Acids Res. 2001.
Abstract
The RegA proteins from the bacteriophage T4 and RB69 are translational repressors that control the expression of multiple phage mRNAs. RegA proteins from the two phages share 78% sequence identity; however, in vivo expression studies have suggested that the RB69 RegA protein binds target RNAs with a higher affinity than T4 RegA protein. To study the RNA binding properties of T4 and RB69 RegA proteins more directly, the binding sites of RB69 RegA protein on synthetic RNAs corresponding to the translation initiation region of two RB69 target genes were mapped by RNase protection assays. These assays revealed that RB69 RegA protein protects nucleotides -9 to -3 (relative to the start codon) on RB69 gene 44, which contains the sequence GAAAAUU. On RB69 gene 45, the protected site (nucleotides -8 to -3) contains a similar purine-rich sequence: GAAAUA. Interestingly, T4 RegA protein protected the same nucleotides on these RNAs. To examine the specificity of RNA binding, quantitative RNA gel shift assays were performed with synthetic RNAs corresponding to recognition elements (REs) in three T4 and three RB69 mRNAs. Comparative gel shift assays demonstrated that RB69 RegA protein has an approximately 7-fold higher affinity for T4 gene 44 RE RNA than T4 RegA protein. RB69 RegA protein also binds RB69 gene 44 RE RNA with a 4-fold higher affinity than T4 RegA protein. On the other hand, T4 RegA exhibited a higher affinity than RB69 RegA protein for RB69 gene 45 RE RNA. With respect to their affinities for cognate RNAs, both RegA proteins exhibited the following hierarchy of affinities: gene 44 > gene 45 > regA. Interestingly, T4 RegA exhibited the highest affinity towards RB69 gene 45 RE RNA, whereas RB69 RegA protein had the highest affinity for T4 gene 44 RE RNA. The helix-loop groove RNA binding motif of T4 RegA protein is fully conserved in RB69 RegA protein. However, homology modeling of the structure of RB69 RegA protein reveals that the divergent residues are clustered in two areas of the surface, and that there are two large areas of high conservation near the helix-loop groove, which may also play a role in RNA binding.
Figures
Figure 1
(A) Sequence of the TIR of RB69 regA on the pAS1-RB69 regA vector. The regA coding region is preceded by the λ cII gene ribosome binding site (underlined) and AUG start codon followed by a 15 nt ORF corresponding to the 3′-end of T4 gene 62 (lower case). Translation initiated at the cII AUG will terminate at the gene 62 stop codon (boxed), and then reinitiate at the regA AUG (italic). (B) Mutations introduced into the TIR of λ _cII_-RB69 regA to increase expression of regA. The ribosome binding site and start codons are in bold.
Figure 2
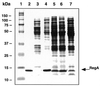
Expression of RB69 and T4 RegA proteins from pAS1 vectors. Total cell extracts from 5 µl of uninduced cell culture (lane 3), 5 µl of nalidixic acid-induced cell cultures expressing T4 RegA protein (lane 4), 10 µl of induced cell cultures expressing RB69 RegA protein from plasmids pG62A (lane 5) (see Fig. 1), pCIIB (lane 6) and WT pAS1-RB69 regA (lane 7), respectively, were applied to a 15% SDS–polyacrylamide gel. Lane 1, molecular weight markers; lane 2, purified T4 RegA protein.
Figure 3
Nucleotide sequence of the translation initiation regions of three RegA-sensitive mRNAs from T4 and RB69 phage. The binding site of T4 RegA protein on T4 gene 44, previously determined by RNase protection assays (3,6), is indicated by shading. The sequences of the oligonucleotides used in this study are indicated by underlining and the initiation codons are in bold type.
Figure 4
Gel mobility shift assay of RB69 RegA protein binding to RE RNAs. 32P-5′-end-labeled 18mer RB69 gene 44 RE RNA (5′-AUGAGGAAAAUUACAUGA-3′) (lanes 1–4), 16mer T4 gene 44 RE RNA (5′-AAUGAGGAAAUUAUG-3′) (lanes 5–7) and 16mer T4 gene 44 G–9U RE RNA (5′-AAU
U
AGGAAAUUAUG-3′) (lanes 8–10) were incubated with cell supernatants containing no RegA protein (lane 2) or 20 nM RB69 RegA protein (lanes 3, 4, 6, 7, 9 and 10). Samples were analyzed by electrophoresis on a native 6% polyacrylamide gel. Gels were dried and then analyzed on a PhosphorImager (see Materials and Methods).
Figure 5
RNase footprint assays of RB69 and T4 RegA protein binding to RB69 gene 44 and RB69 gene 45 TIR RNAs. (A) RB69 gene 44 RE RNA (5′-AUGAGGAAAAUUACAUGA-3′). Lane 1, RNA alone; lanes 2–7, RNA digested with RNase I. Lane 2, RNA digested in the absence of RegA protein; lane 3, RNA digested in the presence of pAS1 cell supernatant (which does not contain RegA protein); lanes 4 and 5, RNA plus cell supernatants containing 20 and 40 nM T4 RegA protein, respectively; lanes 6 and 7, RNA plus cell supernatants containing 20 and 40 nM RB69 RegA protein, respectively. (B) RB69 gene 45 TIR RNA (5′-UGAAAGGAAAUAAAAUGA-3′). Lanes 1–6, RNA digested with RNase I. Lane 1, RNA digested in absence of RegA protein; lane 2, RNA digested in the presence of pAS1 cell supernatant; lanes 3 and 4, RNA plus cell supernatants containing 20 and 40 nM T4 RegA protein, respectively; lanes 5 and 6, RNA plus cell supernatants containing 20 and 40 nM RB69 RegA protein, respectively; lane 7, RNA alone. Reactions in each panel contained 10 nM RNA and 0.01 U/ml of RNase I. RNA fragments were analyzed by electrophoresis on an 8 M urea/TBE gel and detected by PhosphorImager analysis. Nucleotides are numbered relative to the AUG start codon (in bold, above), so that U–4 is 4 nt upstream from the initiation A.
Figure 6
(A) Gel shift binding titration of T4 gene 44 RE RNA with T4 and RB69 RegA proteins. Increasing concentrations of cell supernatants containing T4 or RB69 RegA proteins were incubated with 10 nM 32P-T4 gene 44 RE RNA. RB69 RegA–RNA complexes migrate faster than T4 RegA–RNA complexes, presumably due to RB69 RegA protein’s lower pI value. T4 gene 44 RE RNA: 5′-AAUGAGGAAAUUAUGA-3′. Samples were analyzed as in Figure 4. (B) Competition gel shift assay of T4 and RB69 RegA proteins for T4 gene 44 RE RNA. 32P-T4 gene 44 RE RNA (50 nM) was incubated with cell supernatants containing T4 RegA protein (lane 2), RB69 RegA protein (lane 6) or mixtures of T4 and RB69 proteins (lanes 3–5). Lane 1 in (A and B) contains RNA incubated without cell supernatant. Concentrations of RegA proteins in cell lysates were determined by quantitation of Sypro Orange stained gels using a Molecular Dynamics Storm™ imager.
Figure 7
Comparative gel shift assays of T4 and RB69 RegA proteins binding to RB69 gene 44 (A), RB69 gene 45 (B), T4 gene 45 (C) and RB69 regA (D) RE RNAs. Lanes 1 and 6 in all panels contain RNA incubated without cell supernatants. Lanes 2–5 represent RNA incubated with cell supernatants containing T4 RegA at the indicated concentrations. Lanes 7–10 represent samples containing RB69 RegA at the indicated concentrations. Reactions in (A) and (B) contain 10 nM RB69 gene 44 and gene 45 RE RNAs, respectively; samples in (C) contain 20 nM T4 gene 45 RE RNA; samples in (D) contain 50 nM RB69 regA RE RNA. Binding reactions were analyzed by electrophoresis on native 6% polyacrylamide gels.
Figure 8
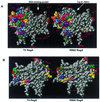
Space-filling models of RB69 and T4 RegA proteins, showing conserved and divergent amino acids. Divergent residues are colored, conserved residues are white. (A) front view, (B) back view, rotated 180°. Note that residues in the RNA binding groove are fully conserved in the two proteins. Arrows indicate the RNA binding pocket and the location of residue Trp81.
Figure 9
RNA gel shift assay of WT and mutant Trp81Ala RB69 RegA proteins. Lanes 1–3 and 4–6, 10 nM T4 and RB69 gene 44 RE RNAs, respectively; lanes 7–9, 50 nM RB69 gene 45 RE RNA. Lanes 2, 3, 5 and 6, 10 nM WT or W81A RB69 RegA proteins was used. Lanes 8 and 9, 50 nM RB69 RegA proteins was used. Samples were analyzed as in Figure 4.
Similar articles
- Post-transcriptional control by bacteriophage T4: mRNA decay and inhibition of translation initiation.
Uzan M, Miller ES. Uzan M, et al. Virol J. 2010 Dec 3;7:360. doi: 10.1186/1743-422X-7-360. Virol J. 2010. PMID: 21129205 Free PMC article. Review. - RNA-protein interactions of the bacteriophage RB69 RegA translational repressor protein.
Jozwik CE, Miller ES. Jozwik CE, et al. Nucleic Acids Symp Ser. 1995;(33):256-7. Nucleic Acids Symp Ser. 1995. PMID: 8643388 - Regions of bacteriophage T4 and RB69 RegA translational repressor proteins that determine RNA-binding specificity.
Jozwik CE, Miller ES. Jozwik CE, et al. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):5053-7. doi: 10.1073/pnas.89.11.5053. Proc Natl Acad Sci U S A. 1992. PMID: 1594613 Free PMC article. - In vitro selection of phage RB69 RegA RNA binding sites yields UAA triplets.
Dean TR, Allen SV, Miller ES. Dean TR, et al. Virology. 2005 May 25;336(1):26-36. doi: 10.1016/j.virol.2005.03.002. Virology. 2005. PMID: 15866068 - Adaptive recognition in RNA complexes with peptides and protein modules.
Patel DJ. Patel DJ. Curr Opin Struct Biol. 1999 Feb;9(1):74-87. doi: 10.1016/s0959-440x(99)80010-4. Curr Opin Struct Biol. 1999. PMID: 10047585 Review.
Cited by
- Jumbo Phages: A Comparative Genomic Overview of Core Functions and Adaptions for Biological Conflicts.
M Iyer L, Anantharaman V, Krishnan A, Burroughs AM, Aravind L. M Iyer L, et al. Viruses. 2021 Jan 5;13(1):63. doi: 10.3390/v13010063. Viruses. 2021. PMID: 33466489 Free PMC article. - Regulation of translation initiation by RNA binding proteins.
Babitzke P, Baker CS, Romeo T. Babitzke P, et al. Annu Rev Microbiol. 2009;63:27-44. doi: 10.1146/annurev.micro.091208.073514. Annu Rev Microbiol. 2009. PMID: 19385727 Free PMC article. Review. - Post-transcriptional control by bacteriophage T4: mRNA decay and inhibition of translation initiation.
Uzan M, Miller ES. Uzan M, et al. Virol J. 2010 Dec 3;7:360. doi: 10.1186/1743-422X-7-360. Virol J. 2010. PMID: 21129205 Free PMC article. Review. - Bacteriophage T4 genome.
Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Rüger W. Miller ES, et al. Microbiol Mol Biol Rev. 2003 Mar;67(1):86-156, table of contents. doi: 10.1128/MMBR.67.1.86-156.2003. Microbiol Mol Biol Rev. 2003. PMID: 12626685 Free PMC article. Review.
References
- Miller E.S., Karam,J.D. and Spicer,E.K. (1994) Translational initiation control: mRNA structure and protein repressors. In Karam,J.D. (ed.), Molecular Biology of Bacteriophage T4. American Society of Microbiology, Washington, DC, pp. 193–205.
- Adari H. and Spicer,E.K. (1986) Translational repression in vitro by the bacteriophage T4 RegA protein. Proteins Struct. Funct. Genet., 1, 116–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources