Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition - PubMed (original) (raw)
Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition
K E Gustin et al. EMBO J. 2001.
Abstract
Infection of eukaryotic cells with lytic RNA viruses results in extensive interactions of viral gene products with macromolecular pathways of the host, ultimately leading to death of the infected cells. We show here that infection of cells with poliovirus results in the cytoplasmic accumulation of a variety of shuttling and non-shuttling nuclear proteins that use multiple nuclear import pathways. In vitro nuclear import assays using semi-permeabilized infected cells confirmed that nuclear import was blocked and demonstrated that docking of nuclear import receptor-cargo complexes at the cytoplasmic face of the nuclear pore complex (NPC) was prevented. Analysis of components of the NPC revealed that two proteins, Nup153 and p62, were proteolyzed during poliovirus infection. These results suggest that the cytoplasmic relocalization of numerous cellular proteins is caused by the inhibition of multiple nuclear import pathways via alterations in NPC composition in poliovirus-infected cells. Blocking of nuclear import points to a novel strategy by which cytoplasmic RNA viruses can evade host immune defenses, by preventing signal transduction to the nucleus.
Figures
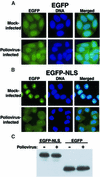
Fig. 1. Intracellular localization of EGFP and EGFP–NLS molecules in uninfected and poliovirus-infected cells. (A) HeLa cells stably expressing EGFP were mock infected or infected with poliovirus as indicated. Cells were processed and examined by fluorescent microscopy at 4.5 h after infection. EGFP fluorescence was visualized using a FITC filter. DNA: Hoechst 33258-stained nuclei were examined with a UV filter. Merged: shows the FITC and Hoechst images merged. (B) HeLa cells stably expressing EGFP–NLS fusion proteins were examined as described in (A). (C) Immunoblot analysis of EGFP–NLS- and EGFP-expressing cell lines. Whole-cell lysates were prepared after 4.5 h of infection from either mock- or poliovirus-infected cells. EGFP was detected by immunoblotting using a polyclonal antibody to GFP (Clontech).
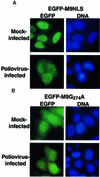
Fig. 2. Intracellular localization of EGFP–M9NLS and EGFP–M9G274A molecules in mock-infected and poliovirus-infected cells. (A) HeLa cells were transiently transfected with plasmid pEGFP-M9NLS and mock infected or infected with poliovirus at 40 h after transfection and examined 4.5 h after infection. Labeling of panels is as described in Figure 1. (B) Same as in (A), except that cells were transfected with plasmid pEGFP-M9G274A.
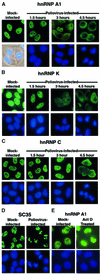
Fig. 3. Intracellular localization of endogenous cellular proteins in uninfected and poliovirus-infected cells. (A) Uninfected (Mock-infected) or HeLa cells infected with poliovirus for 1.5, 3 or 4.5 h were fixed and stained with an antibody directed against hnRNP A1. Top panels show cells examined using a FITC filter, bottom panels show Hoechst 33258 staining of the same field using a UV filter or an overlay of the UV image onto a phase image (Mock-infected). (B) At the times indicated cells were fixed and stained with an antibody directed against hnRNP K. Labeling of the panels is as described for (A). (C) At the times indicated, cells were fixed and stained with a monoclonal antibody directed against hnRNP C. (D) Mock- or poliovirus-infected cells were fixed and stained using a monoclonal antibody specific for SC35. (E) Mock-infected HeLa cells were incubated without or with actinomycin D (5 µg/ml) for 4 h (Act D Treated), then fixed and stained with an antibody directed against hnRNP A1.
Fig. 4. Cell-free nuclear import assays. (A) Uninfected cells (Mock-infected) or cells that had been infected with poliovirus for 4 h (Poliovirus-infected) were permeabilized and used in an in vitro nuclear import assay. Assays were carried out in the presence (+RRL) or absence (–RRL) of rabbit reticulocyte lysate as a source of cytosolic factors. Top panels show GFP using a FITC filter and bottom panels show Hoechst 33258 staining of DNA using a UV filter. All GFP images were acquired using identical exposure times and manipulations in Photoshop 5.0. (B) Same as in (A), except that creatine kinase, creatine phosphate, ATP and GTP were omitted from the reaction. GFP images were acquired using the same exposure time as in (A). (C) Longer exposure of GFP fields in (B).
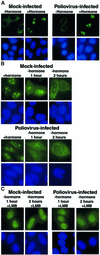
Fig. 5. Intracellular localization of Rev–GC fusion proteins in uninfected and poliovirus-infected cells. (A) HeLa cells were transiently transfected with plasmid pXRGG; after 40 h, cells were either mock infected (Mock-infected) or infected with poliovirus (Poliovirus-infected). Four hours after infection cells were untreated (–hormone) or treated with dexamethasone (+hormone) at 1 µM for 30 min prior to fixation. Top panels show GFP using a FITC filter and bottom panels show Hoechst 33258 staining using a UV filter. (B) Forty hours after transfection with pXRGG, cells were either mock infected or infected with poliovirus and dexamethasone was added. 2.5 h later the cells were either fixed (+hormone), or dexamethasone was removed by washing once with PBS and replacing the medium. Cells were processed for fluorescent microscopy at 1 h (–hormone 1 hour) or 2 h (–hormone 2 hours) following removal of dexamethasone. (C) Same as in (B), except that leptomycin B (5 ng/ml, +LMB) was added following removal of dexamethasone.
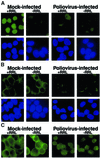
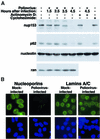
Fig. 6. Alterations in the NPC composition in poliovirus-infected HeLa cells. (A) Whole-cell lysates (50 µg) prepared from cells subjected to various treatments were analyzed by immunoblotting with the indicated antibodies. Actinomycin D: cells were treated for 4.5 h with 5 µg/ml actinomycin D. Cycloheximide: cells were treated with 20 µg/ml cycloheximide for 4.5 h. (B) Indirect immunofluorescence using monoclonal antibodies 414 and SC-7292 to detect nucleoporins and lamins, respectively. Cells were either uninfected (Mock-infected) or infected with poliovirus for 4.5 h (Poliovirus-infected). Top panels show cells examined with a FITC filter, bottom panels show the same fields examined with a UV filter to detect Hoechst 33258 staining. FITC images for a given antibody were acquired with identical exposure times and adjustments.
Similar articles
- Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus.
Gustin KE, Sarnow P. Gustin KE, et al. J Virol. 2002 Sep;76(17):8787-96. doi: 10.1128/jvi.76.17.8787-8796.2002. J Virol. 2002. PMID: 12163599 Free PMC article. - The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins.
Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. Walther TC, et al. EMBO J. 2001 Oct 15;20(20):5703-14. doi: 10.1093/emboj/20.20.5703. EMBO J. 2001. PMID: 11598013 Free PMC article. - Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores.
Belov GA, Lidsky PV, Mikitas OV, Egger D, Lukyanov KA, Bienz K, Agol VI. Belov GA, et al. J Virol. 2004 Sep;78(18):10166-77. doi: 10.1128/JVI.78.18.10166-10177.2004. J Virol. 2004. PMID: 15331749 Free PMC article. - Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: targeting the nuclear pore complex.
Gustin KE. Gustin KE. Virus Res. 2003 Sep;95(1-2):35-44. doi: 10.1016/s0168-1702(03)00165-5. Virus Res. 2003. PMID: 12921994 Free PMC article. Review. - How viruses access the nucleus.
Cohen S, Au S, Panté N. Cohen S, et al. Biochim Biophys Acta. 2011 Sep;1813(9):1634-45. doi: 10.1016/j.bbamcr.2010.12.009. Epub 2010 Dec 15. Biochim Biophys Acta. 2011. PMID: 21167871 Review.
Cited by
- Nipah virus matrix protein: expert hacker of cellular machines.
Watkinson RE, Lee B. Watkinson RE, et al. FEBS Lett. 2016 Aug;590(15):2494-511. doi: 10.1002/1873-3468.12272. Epub 2016 Jul 12. FEBS Lett. 2016. PMID: 27350027 Free PMC article. Review. - Transcriptomic profiling implicates PAF1 in both active and repressive immune regulatory networks.
Kenaston MW, Pham OH, Petit MJ, Shah PS. Kenaston MW, et al. BMC Genomics. 2022 Nov 30;23(1):787. doi: 10.1186/s12864-022-09013-6. BMC Genomics. 2022. PMID: 36451099 Free PMC article. - Picornaviruses and RNA Metabolism: Local and Global Effects of Infection.
Holmes AC, Semler BL. Holmes AC, et al. J Virol. 2019 Oct 15;93(21):e02088-17. doi: 10.1128/JVI.02088-17. Print 2019 Nov 1. J Virol. 2019. PMID: 31413128 Free PMC article. Review. - Human astroviruses: in silico analysis of the untranslated region and putative binding sites of cellular proteins.
De Nova-Ocampo M, Soliman MC, Espinosa-Hernández W, Velez-Del Valle C, Salas-Benito J, Valdés-Flores J, García-Morales L. De Nova-Ocampo M, et al. Mol Biol Rep. 2019 Feb;46(1):1413-1424. doi: 10.1007/s11033-018-4498-8. Epub 2018 Nov 17. Mol Biol Rep. 2019. PMID: 30448895 Free PMC article. Review. - Viral and host proteins involved in picornavirus life cycle.
Lin JY, Chen TC, Weng KF, Chang SC, Chen LL, Shih SR. Lin JY, et al. J Biomed Sci. 2009 Nov 20;16(1):103. doi: 10.1186/1423-0127-16-103. J Biomed Sci. 2009. PMID: 19925687 Free PMC article. Review.
References
- Adam S.A. (1999) Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr. Opin. Cell Biol., 11, 402–406. - PubMed
- Belov G.A., Evstafieva,A.G., Rubtsov,Y.P., Mikitas,O.V., Vartapetian, A.B. and Agol,V.I. (2000) Early alteration of nucleocytoplasmic traffic induced by some RNA viruses. Virology, 275, 244–248. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources