Molecular characterization of cryptosporidium oocysts in samples of raw surface water and wastewater - PubMed (original) (raw)
Molecular characterization of cryptosporidium oocysts in samples of raw surface water and wastewater
L Xiao et al. Appl Environ Microbiol. 2001 Mar.
Abstract
Recent molecular characterizations of Cryptosporidium parasites make it possible to differentiate the human-pathogenic Cryptosporidium parasites from those that do not infect humans and to track the source of Cryptosporidium oocyst contamination in the environment. In this study, we used a small-subunit rRNA-based PCR-restriction fragment length polymorphism (RFLP) technique to detect and characterize Cryptosporidium oocysts in 55 samples of raw surface water collected from several areas in the United States and 49 samples of raw wastewater collected from Milwaukee, Wis. Cryptosporidium parasites were detected in 25 surface water samples and 12 raw wastewater samples. C. parvum human and bovine genotypes were the dominant Cryptosporidium parasites in the surface water samples from sites where there was potential contamination by humans and cattle, whereas C. andersoni was the most common parasite in wastewater. There may be geographic differences in the distribution of Cryptosporidium genotypes in surface water. The PCR-RFLP technique can be a useful alternative method for detection and differentiation of Cryptosporidium parasites in water.
Figures
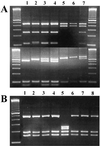
FIG. 1
Genotyping of Cryptosporidium oocysts in water with a SSU rRNA-based PCR-RFLP technique. (A) Differentiation of Cryptosporidium spp. and C. parvum genotypes by digestion of the secondary PCR products with _Ssp_I (upper panel) and _Vsp_I (lower panel). Lane 1, C. parvum human genotype (sample 574); lane 2, C. parvum bovine genotype (sample 5F); lanes 3 and 4, C. parvum human and bovine genotypes (samples 1F and 2F); lane 5, C. andersoni (sample 104); lane 6, C. muris (sample 194); lane 7, C. parvum bovine genotype and C. andersoni (sample 163). (B) Differentiation of C. andersoni from C. muris by digestion of the secondary PCR products with _Dde_I. Lanes 1 through 4 and 6 through 8, C. andersoni (samples 104, 99, 98, 641, 192, 224, and 225); lane 5, C. muris (sample 194).
References
- Chung E, Aldom J E, Carreno R A, Chagla A H, Kostrzynska M, Lee H, Palmateer G, Trevors J T, Unger S, Xu R, De Grandis S A. PCR-based quantitation of Cryptosporidium parvum in municipal water samples. J Microbiol Methods. 1999;38:119–130. - PubMed
- Chung E, Aldom J E, Chagla A H, Kostrzynska M, Lee H, Palmateer G, Trevors J T, Unger S, Degrandis S. Detection of Cryptosporidium parvum oocysts in municipal water samples by the polymerase chain reaction. J Microbiol Methods. 1998;33:171–180.
- Di Giovanni G D, Hashemi F H, Shaw N J, Abrams F A, LeChevallier M W, Abbaszadegan M. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-PCR. Appl Environ Microbiol. 1999;65:3427–3432. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical