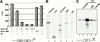Active repression and E2F inhibition by pRB are biochemically distinguishable - PubMed (original) (raw)
Active repression and E2F inhibition by pRB are biochemically distinguishable
J F Ross et al. Genes Dev. 2001.
Abstract
To understand mechanistically how pRB represses transcription, we used a reconstituted transcription assay and compared pRB activity on naked versus chromatin templates. Surprisingly, when pRB was directly recruited to a naked template, no transcriptional repression was observed. However, we observed active repression when the same promoter was assembled into chromatin. Histone deacetylases do not appear to play a role in this observed repression. Further experiments showed repression could occur after preinitiation complex assembly, in contrast with pRB inhibition of E2F, suggesting discrete mechanisms by which pRB directly inhibits an activator such as E2F or actively represses proximally bound transcription factors.
Figures
Figure 1
Gal4-RB directly represses transcription in vivo and retains the ability to inhibit E2F in vitro. (A) Human pRB-deficient C33A cells were transiently cotransfected with the pG5Sp1E4-CAT reporter containing Gal4 and Sp1 binding sites and the indicated plasmids, and CAT assays were performed as described in Materials and Methods. Either no pRB or increasing amounts of pGal4-RB (+, ++, +++, and ++++ indicate 0.25, 1.25, 2.5, and 5 μg of vector, respectively) were included. As controls, full-length pRB (1.25 μg pCMV-RB) and the Gal4 DNA-binding domain (5 μg pSG42Y) also were tested. Representative data from three independent experiments are shown. (B) Silver-stained SDS–polyacrylamide gels of representative purified factors used in the in vitro transcription reactions. (C) In vitro transcription using p(E2F)4B-Gless as a template. E2F, Gal4-RB, and pRB were included as indicated.
Figure 2
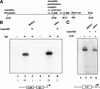
Gal4-RB can repress transcription in vitro on a chromatinized but not a naked template, whereas pRB can inhibit E2F similarly on either template. (A) In vitro transcription using pG5Sp1E4-Gless as a template. Transcription was performed using the indicated factors on a naked (lanes 1_–_7) or chromatin (lanes 8_–_14) template. Five nanograms of Sp1, 20 ng of pRB, 5 ng of Gal4, and 5, 12.5, and 25 ng of Gal4-RB were used per reaction as indicated. The chromatin template was assembled using Drosophila S190 extract. The numbers beneath the lanes represent quantitated relative transcription levels. Basal transcription levels for each panel (lanes 1 and 8) were given unit value. Quantitation was performed as described in Materials and Methods. (B) Transcription reactions were performed as in A, except that reactions included the p(E2F)4B-Gless template, E2F-4/DP-1 (12.5 ng of each subunit per reaction), and 10 ng Gal4-RB. Note that transcription using chromatin results in a lower signal than that of a naked template; hence, autoradiograms displaying transcription from chromatin templates typically required five times longer exposure. (C) In vitro chromatin immunoprecipitations indicate that Gal4-RB does not preclude DNA binding by Sp1. Chromatin was reconstituted with pG5Sp1E4-Gless as a template or mock reconstituted without DNA and incubated with Flag-tagged Sp1, Gal4-RB, or both as indicated above the panel. Proteins and DNA were cross-linked and immunoprecipitated with anti-Flag antibodies as described in Materials and Methods. After reversal of cross-links, samples were electrophoresed and Western blotted sequentially with anti-pRB and anti-Flag antibodies (to detect Sp1). Western blotting with antibodies against Gal4-RB and Sp1 also confirmed that each was present in the indicated reactions before immunoprecipitation (input; data not shown).
Figure 3
Gal4-RB represses transcription on chromatin assembled with recombinant factors. (A) SDS–polyacrylamide gels of the purified factors used to assemble chromatin. The gels containing dNAP1 and ACF were Coomassie- and silver-stained, respectively. (B) Ethidium bromide–stained agarose gels of representative micrococcal nuclease assays to test the assembly of chromatin with S190 extract or recombinant ACF. Increasing amounts of micrococcal nuclease are indicated by the upward sloping triangle. (C) In vitro transcription using chromatin assembled with recombinant factors. Amounts of the indicated factors used were as in Fig. 2. Gal4-RB (12.5 ng) was used in lane 5.
Figure 4
Prior assembly of a preinitiation complex (PIC) does not negate active repression of transcription by Gal4-RB. (A) Diagram of the order of addition experiments. In vitro transcription using (B) pG5Sp1E4-Gless and (C) p(E2F)4BGless templates assembled into chromatin with S190 extract. Sp1 (5 ng) and Gal4-RB (12.5 ng) were included in B, and 12.5 ng of E2F-4/DP-1 and 10 ng of Gal4-RB were added in C as indicated. Gal4-RB was added at the indicated times relative to PIC assembly. Relative transcription levels for B are listed under the lane numbers. Quantitation is relative to basal transcription (lane 1), which was set to unit value as described in the legend to Fig. 2.
Figure 5
Addition of the HDAC inhibitor trichostatin A does not alleviate repression by Gal4-RB. In vitro transcription using chromatin assembled with S190 extract (lanes 1_–_6) or dNAP1 and recombinant ACF (lanes 7_–_12). Sp1 (5 ng), Gal4-RB (12.5 ng), and TSA (0.5 μM) were included as indicated.
Similar articles
- Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein.
Ross JF, Liu X, Dynlacht BD. Ross JF, et al. Mol Cell. 1999 Feb;3(2):195-205. doi: 10.1016/s1097-2765(00)80310-x. Mol Cell. 1999. PMID: 10078202 - Retinoblastoma protein recruits histone deacetylase to repress transcription.
Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Brehm A, et al. Nature. 1998 Feb 5;391(6667):597-601. doi: 10.1038/35404. Nature. 1998. PMID: 9468139 - Dual mechanisms of repression of E2F1 activity by the retinoblastoma gene product.
Zacksenhaus E, Jiang Z, Phillips RA, Gallie BL. Zacksenhaus E, et al. EMBO J. 1996 Nov 1;15(21):5917-27. EMBO J. 1996. PMID: 8918469 Free PMC article. - Molecular mechanisms of E2F-dependent activation and pRB-mediated repression.
Frolov MV, Dyson NJ. Frolov MV, et al. J Cell Sci. 2004 May 1;117(Pt 11):2173-81. doi: 10.1242/jcs.01227. J Cell Sci. 2004. PMID: 15126619 Review. - Activity of the retinoblastoma family proteins, pRB, p107, and p130, during cellular proliferation and differentiation.
Sidle A, Palaty C, Dirks P, Wiggan O, Kiess M, Gill RM, Wong AK, Hamel PA. Sidle A, et al. Crit Rev Biochem Mol Biol. 1996 Jun;31(3):237-71. doi: 10.3109/10409239609106585. Crit Rev Biochem Mol Biol. 1996. PMID: 8817077 Review.
Cited by
- N-(4-Hydroxyphenyl) retinamide potentiated paclitaxel for cell cycle arrest and apoptosis in glioblastoma C6 and RG2 cells.
Janardhanan R, Butler JT, Banik NL, Ray SK. Janardhanan R, et al. Brain Res. 2009 May 1;1268:142-153. doi: 10.1016/j.brainres.2009.02.064. Epub 2009 Mar 10. Brain Res. 2009. PMID: 19285047 Free PMC article. - p110 CUX1 cooperates with E2F transcription factors in the transcriptional activation of cell cycle-regulated genes.
Truscott M, Harada R, Vadnais C, Robert F, Nepveu A. Truscott M, et al. Mol Cell Biol. 2008 May;28(10):3127-38. doi: 10.1128/MCB.02089-07. Epub 2008 Mar 17. Mol Cell Biol. 2008. PMID: 18347061 Free PMC article. - E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex.
Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD. Rayman JB, et al. Genes Dev. 2002 Apr 15;16(8):933-47. doi: 10.1101/gad.969202. Genes Dev. 2002. PMID: 11959842 Free PMC article. - RBP1 family proteins exhibit SUMOylation-dependent transcriptional repression and induce cell growth inhibition reminiscent of senescence.
Binda O, Roy JS, Branton PE. Binda O, et al. Mol Cell Biol. 2006 Mar;26(5):1917-31. doi: 10.1128/MCB.26.5.1917-1931.2006. Mol Cell Biol. 2006. PMID: 16479010 Free PMC article. - Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells.
Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Benhamed M, et al. Nat Cell Biol. 2012 Feb 26;14(3):266-75. doi: 10.1038/ncb2443. Nat Cell Biol. 2012. PMID: 22366686 Free PMC article.
References
- Adnane J, Shao Z, Robbins PD. The retinoblastoma susceptibility gene product represses transcription when directly bound to the promoter. J Biol Chem. 1995;270:8837–8843. - PubMed
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. - PubMed
- Bulger M, Kadonaga JT. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol Genet. 1994;5:241–261.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources